Abstract
The complete mitochondrial genome of the extinct musk ox Bootherium bombifrons is presented for the first time. Phylogenetic analysis supports placement of Bootherium as sister to the living musk ox, Ovibos moschatus, in agreement with morphological taxonomy. SNPs identified in the COI-5p region provide a tool for the identification of Bootherium among material, which is not morphologically diagnosable, for example postcrania, coprolites, and archaeological specimens, and/or lacks precise stratigraphic control, like many from glacial alluvium and in placer mines.
Keywords:
The extant musk ox, Ovibos moschatus, represents only a relict of the Pleistocene diversity of Ovibovini. The helmeted musk ox, Bootherium, was endemic to North America and widespread through Canada and the continental US from mid-Pleistocene until ∼10ka, and by extension is inferred to have occupied a more varied habitat than Ovibos, which is now restricted to the high Nearctic.
Ancient DNA provides a wealth of data to test systematic and spatiotemporal hypotheses. In the case of Ovibos, ancient and modern DNA have been used to reconstruct historical phylogeography and genetic diversity across their Holarctic range (MacPhee et al. Citation2005; MacPhee & Greenwood Citation2007; Campos et al. Citation2010a, Citation2010b). Outcomes of this work indicate that Ovibos were once much more populous (indeed, the fossil record spreads around the entire Arctic), but also that a bottleneck around the time musk oxen became restricted to their current range (∼20ka) has left their extant population genetically depauperate.
Bootherium specimens AMNH-FM 142459, 145489, and 14590 were collected in the mid 20th century from placer mines near Fairbanks, Alaska (N 65.5°, W 148.5°). Target enrichment and IonTorrent sequencing (MYcroarray®, Ann Arbor, MI) with baits from an Ovibos moschatus sequence (Hassanin et al. Citation2009) were conducted on DNA extracted from these three specimens. After assembly in Mira 4.0 (Chevreux et al. Citation1999) and CAP3 (Huang & Madan Citation1999), BLAST + searches followed methods in Kolokotronis et al. (Citation2016). Final assembly used Geneious 8.1.8 (Kearse et al. Citation2012). A complete mitogenome, 16,496 bp long, comprising 13 protein coding genes, two rRNA and 22 tRNA, and control region, was assembled from AMNH-FM 145490 and is on GenBank under accession number KX982584.
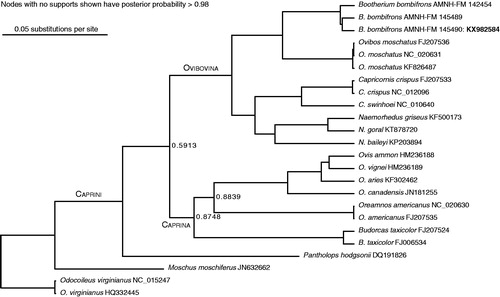
Protein-coding and tRNA genes for the three specimens were aligned against published mitogenomes from 13 caprines and two outgroups. Missing data in the other two Bootherium specimens is in the control region, cytochrome B, and COI. Control region reads were aligned separately from the rest of the genes. Results of the phylogenetic analysis, shown in , support the sister-group relationship of Bootherium to Ovibos. The Bootherium mitogenome shows no major rearrangements. One SNP was identified as diagnosing Bootherium in the barcoding region COI-5p, by comparison to the rest of Caprini, as well as to other bovids including the American bison, with which Bootherium shared much of its range. The divergence time between Bootherium and Ovibos was estimated at ∼250 kyr BP, based on lognormal relaxed molecular clocks, and calibrations from Hassanin et al. (Citation2012). Fossil ovibovine taxonomy centres on a long-established paradigm (Guthrie Citation1992 and references therein) of horn core morphology. Campos et al. (Citation2010b) obtained the first sequences from three extinct musk ox taxa; fragments of the two genes from the Bootherium supported the morphological taxonomy, placing it as a sister genus to Ovibos. However, population dynamics studies on the scale of the Ovibos work are not possible with such limited data. The Bootherium mitogenome will contribute significantly to the available extinct ovibovid molecular information, and in future towards a deeper understanding of extinction dynamics in North America throughout the late Pleistocene.
Figure 1. Maximum posterior probability tree of Caprini, built from complete mitogenomes excluding the control region. Multiple sequence alignment was carried out using Mauve (Darling et al. Citation2004) and ClustalW (Larkin et al. Citation2007). Trees were built in BEAST (Drummond et al. Citation2012) and PAUP (Swofford Citation1991). Model partitions follow the scheme used for Ovis by Sanna et al. (Citation2015).
Acknowledgements
Meng Jin provided access to the AMNH-FM specimens, sampling was supervised by Alana Gishlick; George Amato provided access to molecular lab facilities. Thanks also to Becky Hersch, Anthony Caragiulio, Jon Foox, Zachary Calamari, and John S. S. Denton for laboratory and bioinformatics advice.
Disclosure statement
The authors report no conflicts of interest.
Funding
This work was supported by the National Science Foundation under Grant, 10.13039/100000001 [DGE-16-44869]; and Lamont-Doherty Earth Observatory under a grant from the Chevron Student Initiative Fund.
References
- Campos PF, Willerslev E, Sher A, Orlando L, Axelsson E, Tikhonov A, Aaris-Sorensen K, Greenwood AD, Kahlke RD, Kosintsev P, et al. 2010a. Ancient DNA analyses exclude humans as the driving force behind late Pleistocene musk ox (Ovibos moschatus) population dynamics. Proc Natl Acad Sci USA. 107:5675–5680.
- Campos PF, Sher A, Mead JI, Tikhonov A, Buckley M, Collins M, Willerslev E, Gilbert MTP. 2010b. Clarification of the taxonomic relationship of the extant and extinct ovibovids, Ovibos, Praeovibos, Euceratherium and Bootherium. Quat Sci Rev. 29:2123–2130.
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. In: Proceedings of the GCB '99: German Conference on Bioinformatics. 4-6 October 1999, Hannover, Germany.
- Darling ACE, Mau R, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973.
- Guthrie RD. 1992. New paleoecological and paleoethological information on the extinct helmeted muskoxen from Alaska. Ann Zool Fennici. 28:175–186.
- Hassanin A, Delsuc F, Ropiquet A, Hammer C, Jansen van Vuuren B, Matthee C, Ruiz-Garcia M, Catzefliz F, Areskoug V, Nguyen TT, Couloux A. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol. 335:32–50.
- Hassanin A, Ropiquet A, Couloux A, Cruaud C. 2009. Evolution of the mitochondrial genome in mammal living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol. 68:293–310.
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kolokotronis S-O, Foox J, Rosenfeld JE, Brugler MR, Reeves D, Benoit JB, Booth W, Robison G, Steffen M, Sakas Z, et al. 2016. The mitogenome of the bed bug Cimex lectularius (Hemiptera: Cimicidae). Mitochondrial DNA B Resour. 1:425–427.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948.
- MacPhee RDE, Greenwood AD. 2007. Continuity and change in the extinction dynamics of late Quaternary muskox (Ovibos): genetic and radiometric evidence. Bull Carnegie Mus Nat Hist. 39:203–212.
- MacPhee RDE, Tikhonov AN, Mol D, Greenwood AD. 2005. Late quaternary loss of genetic diversity in muskox (Ovibos). BMC Evol Biol. 5:1471–2148.
- Sanna D, Barbato M, Hadjisterkotis E, Cossu P, Decandia L, Trova S, Pirastru M, Leone GG, Naitana S, Francalacci P, et al. 2015. The first mitogenome of the Cyprus mouflon (Ovis gmelini ophion): new insights into the phylogeny of the genus Ovis. PLoS One. 10:e0144257.
- Swofford DL. 1991. PAUP: Phylogenetic Analysis Using Parsimony, Version 3.1 Computer program distributed by the Illinois Natural History Survey, Champaign, Illinois.
