Abstract
The first complete mitochondrial genome sequence of parthenogenetic Caucasian rock lizard Darevskia unisexualis (Lacertidae family) is determined by hybrid assembly with Illumina HiSeq and PacBio RS II platforms. The circular 21.4 kbp mitogenome contains 13 protein-coding genes, 12S and 16S rRNA genes, 20 tRNAs, two pseudogenized tRNAs, and one long tandem repeats with 4.1 kbp length formed by 59 bp monomer repeated x70.6 times located before control region. This finding represents a new example of mitogenome variation in lizards of hybrid origin, providing fundamental data for following study of a unique hybridization system formed by parthenogenetic and bisexual species in the mountain steppe of central Armenia.
Parthenogenetic lizard species represent a unique model of organisms for study of genetic and ecological bases of hybridogeneous speciation. Darevskia unisexualis is one of the seven parthenogenetic species originated from the interspecific hybridization of D. raddei nairensis (maternal species) and D. valentini (paternal species), which belong to different clades (Moritz et al. Citation1992; Murphy et al. Citation2000; Fujita & Moritz Citation2010).
Mitochondrial genomes of Squamata often have unusual and complicated structure (Amer and Kumazawa Citation2005; Dong & Kumazawa Citation2005; Amer & Kumazawa Citation2007; Fujita et al. Citation2007; Kumazawa et al. Citation2014). In this article we report a new kind of rearrangement in mitochondrial genome of asexual lizard Darevskia unisexualis: a long tandem repeat with 59 bp monomer located before the control region (Darevsky et al. Citation1985; Moritz et al. Citation1992; Murphy et al. Citation2000; Fujita & Moritz Citation2010).
DNA samples extracted from blood of D. unisexualis obtained previously between 1997 and 2006 from lizards of Armenian population Takyarlu (Artavaz, 40°37'20.2”N 44°34’51.4”E) were delivered to the Axeq/Macrogen (Korea) for genomic library preparation and sequencing with Illumina HiSeq 2000 and PacBio RS II.
To overcome complexity of control region assembly we used approach based on reads extraction with following greedy assembly previously successfully adopted in Solenodon paradoxus mitogenome assembly (Brandt et al. Citation2016). A subset of Illumina reads having common kmers with Podarcis siculus mitogenome (NC_011609; Podnar et al. Citation2009) was extracted with Cookiecutter software (Starostina et al. Citation2015), mapped back to the reference mitogenome with bowtie2 (Langmead & Salzberg Citation2012), and assembled into consensuses sequence, that was used as query for alignment against PacBio reads with Blasr aligner (Chaisson & Tesler Citation2012). Cookiecutter’s kmer database was updated from 215 successfully found PacBio reads. Updated kmer database was used for accurate read extraction from Illumina HiSeq raw data. After that Illumina reads were mapped back to PacBio reads and assembled to the final sequence. The consistency of assembly was verified: (1) by consistency with PacBio reads that were mapped to it with MAFFT tool (Katoh & Standley Citation2013); (2) by consistency with previously published restriction map (Moritz et al. Citation1992) that has a perfect match with assembled mitogenome except two inverted restriction sites; (3) consistency of Illumina reads coverage. The assembled genome was annotated with MITOS WebServer (Bernt et al. Citation2013) with additional checking for predicted tRNA and protein-coding genes.
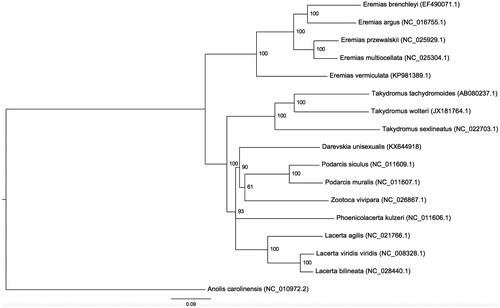
The circular assembled mitogenome size is 21,433 bp, with 13 predicted protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 20 tRNAs, two pseudogenized tRNAs, and one complex tandem repeats with 59 bp monomer repeated x70.6 times and located before control region. MITOS WebServer predict second ND4 in 17202–18491 regions inside found tandem repeat, however, verification by Blast alignment did not find any significant match in this region. The validation of phylogenetic position is shown in .
Figure 1. Molecular phylogeny of Daresvskia unisexualis and other Lacertidae species based on all mitochondrial protein-coding genes and rRNAs. Each of alignments was performed separately by MAFFT v7.187 (Katoh & Standley Citation2013), then alignments were concatenated. Tree was reconstructed by RAxML v8.0.22 (Stamatakis Citation2014) with 10,000 bootstrap replicates. All nodes with support lower than 50 were removed using ETE toolkit (Huerta-Cepas et al. Citation2010) and final tree was drawn by FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Unusual mitochondrial genome of parthenogenetic lizards containing long tandem repeat raises questions about stability and evolution of mtDNA after recent hybridization events. Moreover, its features provide unique opportunities to study mtDNA evolution not only in Squamata taxon, but in all animals.
Data availability
D. unisexualis mitogenome assembly is available from Genebank with accession number KX644918.
Disclosure statement
The authors declare that they have no conflicts of interest concerning this article.
References
- Amer SA, Kumazawa Y. 2005. Mitochondrial genome of Pogona vitticepes (Reptilia; Agamidae): control region duplication and the origin of Australasian agamids. Gene. 346:249–256.
- Amer SA, Kumazawa Y. 2007. The mitochondrial genome of the lizard Calotes versicolor and a novel gene inversion in South Asian draconine agamids. Mol Biol Evol. 24:1330–1339.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Brandt AL, Grigorev K, Afanador-Hernández YM, Paulino LA, Murphy WJ, Núñez A, Komissarov A, Brandt JR, Dobrynin P, Hernández-Martich JD, María R. 2016. Mitogenomic sequences support a north–south subspecies subdivision within Solenodon paradoxus. Mitochondrial DNA Part A. 14:1–9.
- Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinform. 13:238.
- Darevsky IS, Kupriyanova LA, Uzzell T. 1985. Parthenogenesis in reptiles. In: Gans C, Billett DF, editors. Biology of Reptilia. vol. 15. New York: John Wiley & Sons; p. 413–526.
- Dong S, Kumazawa Y. 2005. Complete mitochondrial DNA sequences of six snakes: phylogenetic relationships and molecular evolution of genomic features. J Mol Evol. 61:12–22.
- Fujita MK, Boore JL, Moritz C. 2007. Multiple origins and rapid evolution of duplicated mitochondrial genes in parthenogenetic geckos (Heteronotia binoei; Squamata, Gekkonidae). Mol Biol Evol. 24:2775–2786.
- Fujita MK, Moritz C. 2010. Origin and evolution of parthenogenetic genomes in lizards: current state and future directions. Cytogenet Genome Res. 127:261–272.
- Huerta-Cepas J, Dopazo J, Gabaldón T. 2010. ETE: a python environment for tree exploration. BMC Bioinform. 11:1.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumazawa Y, Miura S, Yamada C, Hashiguchi Y. 2014. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genom. 15:1.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Moritz C, Uzzell T, Spolsky C, Hotz H, Darevsky I, Kupriyanova L, Danielyan F. 1992. The material ancestry and approximate age of parthenogenetic species of Caucasian rock lizards (Lacerta: Lacertidae). Genetica. 87:53–62.
- Murphy RW, Fu J, Macculloch RD, Darevsky IS, Kupriyanova LA. 2000. A fine line between sex and unisexuality: the phylogenetic constraints on parthenogenesis in lacertid lizards. Zool J Linn Soc. 130:527–549.
- Podnar M, Pinsker W, Mayer W. 2009. Complete mitochondrial genomes of three lizard species and the systematic position of the Lacertidae (Squamata). J Zool Syst Evol Res. 47:35–41.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Starostina E, Tamazian G, Dobrynin P, O’Brien SJ, Komissarov A. 2015. Cookiecutter: a tool for kmer-based read filtering and extraction. bioRxiv. Available from: http://dx.doi.org/10.1101/024679.
