Abstract
The complete mitochondrial genomes of two species of the North American collared lemmings were obtained by using PCR amplification and capillary sequencing (GenBank accession nos. KX712239 and KX683880). The collared lemming mitochondrial genomes are 16,341 and 16,338 bp long and show the gene order, contents and gene strand asymmetry typical for mammals. The mitogenome sequences provide an important genomic resource for the collared lemmings, which are model study species in Arctic genetic diversity and biogeographic history.
The North American collared lemmings, Dicrostonyx groenlandicus and D. hudsonius, key species of the Arctic communities, inhabit treeless tundra in Alaska and North Western Canada. Mitochondrial DNA data available for these species are limited to fragments of single gene or single region sequences (Fedorov & Goropashnaya Citation1999; Ehrich et al. Citation2000; Fedorov & Stenseth Citation2002). The North American phylogeography based on partial sequences of the mitochondrial cytochrome b gene and control region revealed relatively low genetic variation in the collared lemmings and provided limited information content for inferring demographic history (Fedorov & Stenseth Citation2002). Access to the complete mitogenomes of these species will facilitate high resolution population genetics analyses.
We present the first complete mitochondrial genome sequences of the North American collared lemmings, Dicrostonyx groenlandicus (NCBI GenBank database accession no. KX712239) and Dicrostonyx hudsonius (GenBank accession no. KX683880). The mitogenomes sequenced belong to D. groenlandicus collected in Alaska (coordinates: 68.338/−158.727) and D. hudsonius from Ungava Peninsula, Canada (coordinates: 62.333/−73.667), and voucher specimens (UAM:Mamm:56332 and UAM:Mamm:57927) were deposited to the Mammal Collection, Museum of the North, University of Alaska Fairbanks. The total genomic DNA was extracted from liver tissue, the mitogenome was amplified by polymerase chain reaction in two overlapping amplicons and capillary sequenced by primer walking (the primer sequences are available on request). The collared lemming mitogenomes are 16,341 bp and 16,338 bp long, with a base composition on the heavy strand of A – 33%, C – 27%, G – 13%, T – 27%, and consist of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and a control region. The order and direction of these genes were identical to those of other Metazoa (Gissi et al. Citation2008). Most of the genes encoded by the heavy strand, except for nad6 and eight tRNA genes encoded by the light strand. All tRNAs were predicted to fold into typical cloverleaf secondary structures. Of the 13 protein-coding genes, three (nad1, cox3 and nad4) showed an incomplete stop codon which is completed by the addition of 3' A residues to the mRNA.
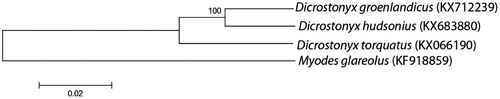
Phylogenetic tree () constructed with mitochondrial genome sequences excluding hypervariable control regions shows that the North American D. groenlandicus and D. hudsonius form well-supported monophyletic group relatively the Eurasian D. torquatus as it was shown by phylogeny based on the cytochrome b gene (Fedorov & Goropashnaya Citation1999). This implies one colonization event from Eurasia through the Bering Land Bridge to North America followed by vicariant separation by the Pleistocene ice sheets. Monophyly of the North American species does not support the hypothesis that the morphologically primitive D. hudsonius is a relict of an earlier colonization of North America from Eurasia while D. groenlandicus represents second colonization event (Guilday Citation1963).
Figure 1. Neighbour-joining tree based on Tamura-Nei nucleotide distances showing phylogenetic relationships among the Eurasian D. torquatus and North American D. groenlandicus and D. hudsonius. The mtgenome of vole Myodes glareolus was included as outgroup. Total of 15,442 bp were used in the alignment excluding hypervariable control region. Number on the node indicates bootstrap support and numbers in brackets correspond to GenBank accession numbers for complete mitochondrial genomes.
The complete mitogenome sequences reported here provide a new genomic resource for the model species in studies of Arctic genetic diversity and biotic history.
Acknowledgements
We thank L. E. Olson, Mammal Collection Museum of the North University of Alaska Fairbanks for providing tissue samples.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Ehrich D, Fedorov VB, Stenseth NC, Krebs CJ, Kenney A. 2000. Phylogeography and mitochondrial DNA diversity in North American collared lemmings Dicrostonyx groenlandicus. Mol Ecol. 9:329–337.
- Fedorov VB, Goropashnaya AV. 1999. The importance of Ice Ages in diversification of Arctic collared lemmings (Dicrostonyx): evidence from the mitochondrial cytochrome B region. Hereditas. 130:301–307.
- Fedorov VB, Stenseth NC. 2002. Multiple glacial refugia in the North American Arctic: inference from phylogeography of the collared lemming (Dicrostonyx groenlandicus). Proc Biol Sci. 269:2071–2078.
- Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 101:301–320.
- Guilday JE. 1963. Pleistocene zoogeography of the lemming, (Dicrostonyx). Evolution. 17:194–197.
