Abstract
The complete mitochondrial genome of Salamandra salamandra was reconstructed using bycatch sequences from an RNAseq library generated from muscle tissue. This study shows the potential of using data originally produced for transcriptome assembly to additionally generate complete mitochondrial genomes. The resulting mitogenome was circular, consisted of 16,331 bp and followed the standard vertebrate gene order. Subsequent analysis of the current mitochondrial genome sequence for the species as listed on GenBank highlighted a species misidentification.
Keywords:
The Salamandra genus includes six species (S. algira, S. atra, S. corsica, S. infraimmaculata, S. lanzai and S. salamandra) of which the European fire salamander (Salamandra salamandra, Linnaeus 1758) is the most widely spread and is distributed across much of Central and Southern Europe (Steinfartz et al. Citation2000; Vences et al. Citation2014). It exhibits high colour and morphological polymorphism resulting in the identification of several subspecies (Thiesmeier & Grossenbacher Citation2004; Beukema et al. Citation2016), and it shows remarkable variation in reproductive mode (Buckley et al. Citation2007; Velo-Antón et al. Citation2015).
Although, GenBank currently holds a mitochondrial genome identified as S. salamandra (EU880331), it was already observed by Ben Hassine et al. (Citation2016) that based on sampling locality (Kordestan Province, Iran) this specimen is most likely a misidentification and should instead be classified as S. infraimmaculata. Using RNAseq data collected for transcriptome assembly, we reconstructed the mitogenome of one individual from within the known S. salamandra range in Northern Portugal (Mindelo; 41.326768, −8.733695). Total RNA was extracted from 35 g of muscle tissue using the RNeasy mini kit (Qiagen, Hilden, Germany). The library was prepared following the standard TruSeq protocol (Illumina) and sequenced on a HiSeq lane for 2x126bp sequencing resulting in over 25 million paired end reads. Illumina adapters were removed using cutadapt version 1.9 (Martin Citation2011) and poly-A-tails were trimmed and duplicate sequences were removed using prinseq 0.20.4 (Schmieder & Edwards Citation2011).
In order to make sure the reference would not bias our results, we used an iteration-based method using a 1422bp Sanger sequence from cytochrome b from the same population as a starting point. Prior to the iterative assembly as performed in Geneious 9.1.2 (Kearse et al. Citation2012), reads were paired using pear 0.9.6 (Zhang et al. Citation2014) and using a custom awk scripts only paired reads of above 200 bps were retained. Following 76 iterations the resulting contig was circular and the iterations were stopped. Additionally, phylogenetic analyses including all Salamandra species and most subspecies were conducted using the same dataset and parameters as was used in the most recent phylogeny of this genus (Vences et al. Citation2014).
The resulting mitochondrial genome was 16,331 bp long, had a GC-content of 36.8% and followed the standard vertebrate gene order. It had 1153 SNPs and 19 indels compared to the current GenBank reference for S. salamandra (the putative S. infraimmaculata) and is over 7% divergent. Bayesian inference analysis of the mtDNA dataset supports monophyly at species level and shows that the current mitogenome on GenBank (EU80331) groups with the S. infraimmaculata clade, whereas the reconstructed mitogenome from this study is embedded within the S. salamandra clade, with the S. s. gallaica specimen as its closest relative ().
Figure 1. Phylogenetic tree of representative species and most subspecies from the Salamandra genus using Lyciasalamandra as an outgroup. Black circles indicate well-supported nodes (posterior probabilities >0.95). Highlighted by boxes are the original GenBank entry (EU880331) for Salamandra salamandra which groups with Salamandra infraimmaculata and the sample used for this study Salamandra salamandra gallaica (KX094979) which groups with the correct clade.
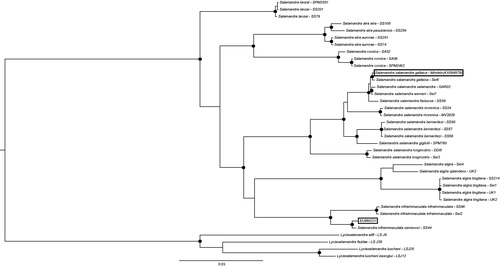
Taking sequence as well as location information content into account we classify EU880331 as S. infraimmaculata. The Salamandra salamandra mitogenome resulting from this paper has been submitted to NCBI under ascension number KX094979. This is the first published mitogenome for this species and only the second complete mitogenome for this well-studied genus. It also highlights the potential of using RNAseq bycatch data to additionally reconstruct complete mitochondrial genomes.
Acknowledgements
We thank Sandra Afonso for help with laboratory work.
Disclosure statement
The authors report no declarations of interest.
References
- Ben Hassine J, Gutiérrez-Rodríguez J, Escoriza D, Martínez-Solano I. 2016. Inferring the roles of vicariance, climate and topography in population differentiation in Salamandra algira (Caudata, Salamandridae). J Zool Syst Evol Res. 54:116–126.
- Beukema W, Nicieza AG, Lourenço A, Velo-Antón G. 2016. Colour polymorphism in Salamandra salamandra (Amphibia: Urodela), revealed by a lack of genetic and environmental differentiation between distinct phenotypes. J Zool Syst Evol Res. 54:127–136.
- Buckley D, Alcobendas M, García-París M, Wake MH. 2007. Heterochrony, cannibalism, and the evolution of viviparity in Salamandra salamandra. Evol Dev. 9:105–115.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27:863–864.
- Steinfartz S, Veith M, Tautz D. 2000. Mitochondrial sequence analysis of Salamandra taxa suggests old splits of major lineages and postglacial recolonizations of Central Europe from distinct source populations of Salamandra salamandra. Mol Ecol. 9:397–410.
- Thiesmeier B, Grossenbacher K. 2004. Salamandra salamandra (Linnaeus, 1758)–Feuersalamander. Handb Reptil Amphib Eur Schwanzlurche. IIB:1059–1132.
- Velo-Antón G, Santos X, Sanmartín-Villar I, Cordero-Rivera A, Buckley D. 2015. Intraspecific variation in clutch size and maternal investment in pueriparous and larviparous Salamandra salamandra females. Evol Ecol. 29:185–204.
- Vences M, Sanchez E, Hauswaldt JS, Eikelmann D, Rodríguez A, Carranza S, Donaire D, Gehara M, Helfer V, Lötters S, et al. 2014. Nuclear and mitochondrial multilocus phylogeny and survey of alkaloid content in true salamanders of the genus Salamandra (Salamandridae). Mol Phylogenet Evol. 73:208–216.
- Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 30:614–620.
