Abstract
Androglossini is one of four tribes recognized within a neotropical parrot subfamily Arinae. The tribe includes 10 genera of which Pionus is represented by eight species. However, its evolutionary diversification and relationship with other Androglossini members are still unclear. Depending on studied molecular markers, Pionus is closely related with Amazona genus or two monotypic genera Alipiopsitta and Graydidascalus or the clade in which Amazona genus is sister to Alipiopsitta and Graydidascalus. Therefore, we sequenced Pionus menstruus menstruus mitogenome to gain molecular data appropriate for future studies to resolve these discrepancies obtained in various phylogenetic analyses published so far.
Neotropical parrots are classified into Arinae subfamily, in which four tribes were recognized (Schodde et al. Citation2013). One of them is Androglossini, which includes 10 genera with Pionus represented by eight species: chalcopterus, fuscus, maximiliani, menstruus, senilis, seniloides, sordidus, and tumultuosus (Forshaw Citation2010). Four of them are monotypic but chalcopterus, menstruus, maximiliani, and sordidus are divided into two, three, four, and six subspecies, respectively (Ribas et al. Citation2007). Most of Pionus taxa are distributed in South America except for Central American P. senilis as well as P. menstruus rubrigularis, which inhabit a region from Costa Rica to Ecuador.
So far, Ribas et al. (Citation2007) presented the most comprehensive study of Pionus including 257 specimens of all known subspecies. Their analyses showed the influence of Andes uplift and Pleistocene climatic oscillations on phylogenetic differentiation and biogeographic disjunctions of montane lineages and taxa inhabiting lowland dry and wet forests. However, their molecular analyses did not completely clarify evolutionary history of this genus. Different phylogenetic methods based on two concatenated mitochondrial sequences from cytochrome b and NADH dehydrogenase 2 produced contradictory tree topologies and many internal branches obtained weak or no statistical support. One of these inconsistencies concerns the placement of monotypic species P. fuscus, which is located basally to other Pionus species or within the tree.
Moreover, the evolutionary relationship of the whole genus Pionus with other Androglossini members also raises a lot of doubts based on the phylogenies published so far. Depending on the usage of molecular markers, Pionus is related with two monotypic genera, Alipiopsitta and Graydidascalus (Schweizer et al. Citation2014; Urantówka et al. Citation2014) or Amazona genus (Kirchman et al. Citation2012) or the clade in which Amazona genus is sister to both Alipiopsitta and Graydidascalus genera (Ribas et al. Citation2007) or Graydidascalus (Schirtzinger et al. Citation2012).
These controversies imply that more molecular data are required to reconstruct precise phylogeny of Pionus. Since it was shown that complete mitochondrial genomes can provide useful information for evolutionary studies of many taxa (Nabholz et al. Citation2013) and no complete Pionus mitogenome is available, we sequenced the mitogenome from P. menstruus menstruus to gain appropriate molecular data for future examination of Pionus diversification and its relationship to other Androglossini members. This subspecies is one of three from P. menstruus species and has the most widespread distribution in Amazonian region. The range of isolated reichenowi subspecies is limited to southeastern Brazil’s South Atlantic coast, whereas rubrigularis also has a disjunctive population in northwestern South America.
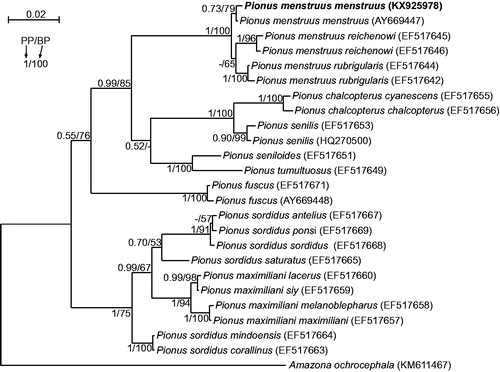
The sequence of P. menstruus menstruus genome with the length of 18,545 bp was deposited in GenBank under the accession number KX925978. Although morphology of the analyzed specimen (Polish captive bird) was absolutely typical for other P. menstruus menstruus individuals, we proved its taxonomic affiliation in phylogenetic analyses of nd2 sequences including all available Pionus taxa with confirmed geographic origins. The obtained tree () revealed that the analyzed individual grouped significantly with another representative of its subspecies. This group was sister to the clade with subspecies rubrigularis and reichenowi.
Figure 1. The phylogenetic tree obtained in MrBayes for nd2 gene indicating that the studied individual (bolded) belongs to P. menstruus menstruus. The parrot is kept in aviculture and its blood sample from which DNA was isolated is available in the laboratory at the Department of Genetics in Wroclaw University of Environmental and Life Sciences under the number ADUAKPM02. All P. menstruus subspecies create a significant monophyletic clade but P. sordidus subspecies are separated by the clade of P. maximiliani. It implies that P. sordidus is a polyphyletic taxon and should be revised. Values at nodes, in the order shown, indicate posterior probabilities found in MrBayes (PP) and bootstrap percentages calculated in TreeFinder (BP). In the MrBayes (Ronquist et al. Citation2012) analysis, separate mixed substitution models were assumed for three codon positions with information about heterogeneity rate across sites as proposed by PartitionFinder (Lanfear et al. Citation2012). We applied two independent runs, each using four Markov chains. Trees were sampled every 100 generations for 10,000,000 generations. After obtaining the convergence, trees from the last 4,717,000 generations were collected to compute the posterior consensus. In the case of TreeFinder (Jobb et al. Citation2004), the separate substitution models were selected for three codon positions according to Propose Model module in this program, and 1000 replicates were assumed in the bootstrap analysis. The posterior probabilities <0.5 and bootstrap percentages <50 were marked by a dash ‘-’.
Disclosure statement
The authors report no conflicts of interest and alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Forshaw JM. 2010. Parrots of the world. London: A & C Black Publishers Ltd; p. 328.
- Jobb G, von Haeseler A, Strimmer K. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 4:18.
- Kirchman JJ, Schirtzinger EE, Wright TF. 2012. Phylogenetic relationships of the extinct Carolina Parakeet (Conuropsis Carolinensis) inferred from DNA sequence data. Auk. 129:197–204.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Nabholz B, Uwimana N, Lartillot N. 2013. Reconstructing the phylogenetic history of long-term effective population size and life-history traits using patterns of amino acid replacement in mitochondrial genomes of mammals and birds. Genome Biol Evol. 5:1273–1290.
- Ribas CC, Moyle RG, Miyaki CY, Cracraft J. 2007. The assembly of montane biotas: linking Andean tectonics and climatic oscillations to independent regimes of diversification in Pionus parrots. Proc Biol Sci. 274:2399–2408.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Schirtzinger EE, Tavares ES, Gonzales LA, Eberhard JR, Miyaki CY, Sanchez JJ, Hernandez A, Mueller H, Graves GR, Fleischer RC, et al. 2012. Multiple independent origins of mitochondrial control region duplications in the order Psittaciformes. Mol Phylogenet Evol. 64:342–356.
- Schodde R, Remsen JV, Schirtzinger EE, Joseph L, Wright TF. 2013. Higher classification of New World parrots (Psittaciformes; Arinae), with diagnoses of tribes. Zootaxa. 3691:591–596.
- Schweizer M, Hertwig ST, Seehausen O, Ebach M. 2014. Diversity versus disparity and the role of ecological opportunity in a continental bird radiation. J Biogeography. 41:1301–1312.
- Urantówka AD, Mackiewicz P, Strzała T. 2014. Phylogeny of Amazona barbadensis and the Yellow-headed Amazon complex (Aves: Psittacidae): a new look at South American parrot evolution. PLoS One. 9:e97228.
