Abstract
The Korean flying fish, Cheilopogon doederleinii, is a marine fish species belonging to the family Exocoetidae. In this study, I report for the first time the sequencing and assembly of the complete mitochondrial genome of C. doederleinii. The complete mitochondrial genome is 16,525 bp long and includes 13 protein-coding, 22 tRNA, and 2 rRNA genes. It has the typical vertebrate mitochondrial gene arrangement. Phylogenetic analysis using mitochondrial genomes of 10 species showed that C. doederleinii is clustered with C. arcticeps and grouped with the other Exocoetidae species. This mitochondrial genome provides potentially important resources for addressing taxonomic issues and studying molecular evolution.
The Korean flying fish, Cheilopogon doederleinii, is a marine fish species belonging to the family Exocoetidae. This species is widely distributed in the western Pacific Ocean and is commercially fished in Korea, Japan, and China (Kim et al. Citation2005). To date, there has been very little molecular and genetic research on this species. To the best of my knowledge, this is the first study to determine the complete mitochondrial genome of C. doederleinii and to analyze the phylogenetic relationship of this species among Beloniformes fishes.
The C. doederleinii specimen collected from the Jeju Island in Korea (33.13N, 126.14E) was deposited in the National Marine Biodiversity Institute of Korea (Specimen No. MABIK PI00045941). The germeline stem cells of this species were cryopreserved in the National Institute of Biological Resources (Incheon, Korea) with a slight modification to previously reported methods (Lee et al. Citation2013; Lee et al. Citation2016). The mitochondrial genome was amplified in its entirety using a long PCR technique (Cheng et al. Citation1994). I used fish-versatile PCR primers in various combinations to amplify contiguous, overlapping segments of the entire mitogenome. All the experiments were carried out following previously described methods (Miya & Nishida Citation1999).
The complete mitochondrial genome of C. doederleinii (GenBank accession no. AP017897) is 16,525 bp long and includes 13 protein-coding, 22 tRNA, and 2 rRNA genes.
The ND6 gene and eight tRNA genes are encoded on the light-strand. The overall base composition of the heavy-strand is 29.22% of A, 27.15% of C, 16.29% of G, and 27.31% of T. As in mitogenomes of other vertebrates (Saccone et al. Citation1999), the AT content is higher than the GC content. All tRNA genes can fold into a typical cloverleaf structure and are 65–74 bp long. The 12S rRNA (944 bp) and 16S rRNA genes (1686 bp) are located between tRNAPhe and tRNAVal and between rRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 12 start with ATG; the exception being COI, which starts with GTG. Seven of the 13 protein-coding genes terminate with incomplete stop codons, T–– (ND2, COII, ND3, ND4, and Cytb) and TA– (ATP6 and COIII), whereas the remaining six genes ended with complete stop codons (TAG and TAA). A control region (866 bp) is located between tRNAPro and tRNAPhe.
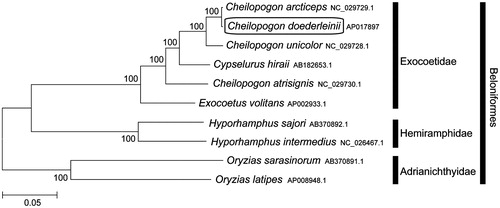
Phylogenetic trees were constructed (maximum likelihood) with 1000 replicates using MEGA 7.0 software (MEGA, PA) (Kumar et al. Citation2016) for the newly sequenced mitogenome and a further nine complete mitogenome sequences downloaded from the National Center for Biotechnology Information. I confirmed that C. doederleinii is clustered with C. arcticeps (Chou et al. Citation2016) and grouped with the other Exocoetidae species (). This mitochondrial genome provides potentially important resources for addressing taxonomic issues and studying molecular evolution.
Disclosure statement
The author reports no conflict of interests. The author alone is responsible for the content and writing of the paper.
Funding
This study was supported by the National Institute of Biological Resources under grant number NIBR201612107.
References
- Cheng S, Higuchi R, Stoneking M. 1994. Complete mitochondrial genome amplification. Nat Genet. 7:350–351.
- Chou CE, Wang FY, Chang SK, Chang HW. 2016. Complete mitogenome of three flyingfishes Cheilopogon unicolor, Cheilopogon arcticeps and Cheilopogon atrisignis (Teleostei: Exocoetidae). Mitochondrial DNA Part B. 1:142–143.
- Kim IS, Choi Y, Lee CL, Lee YJ, Kim BJ, Kim JH. 2005. Illustrated book of Korean fishes. Seoul, Korea: Kyo-Hak Press.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee S, Iwasaki Y, Shikina S, Yoshizaki G. 2013. Generation of functional eggs and sperm from cryopreserved whole testes. Proc Natl Acad Sci USA. 110:1640–1645.
- Lee S, Katayama N, Yoshizaki G. 2016. Generation of juvenile rainbow trout derived from cryopreserved whole ovaries by intraperitoneal transplantation of ovarian germ cells. Biochem Biophys Res Commun. 478:1478–1483.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1:416–426.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.