Abstract
The southern brook lamprey (Ichthyomyzon gagei) is a non-parasitic lamprey endemic to the southeastern US. Here, we report the complete mitogenome of this basal vertebrate and found its genomic organization to be similar to that of other reported lamprey mitogenomes. Nucleotide sequence identities for individual proteins range from 90% to 94% when compared with the congeneric species I. fossor and I. unicuspis. Finally, phylogenetic analysis revealed I. gagei to be sister to these other species of Ichthyomyzon. These genomic data provide a baseline for future investigations regarding the molecular evolution of basal vertebrates.
The lampreys are important model organisms in evolutionary studies because of their peculiar life histories and their retention of many features reminiscent of the earliest vertebrates (Osorio and & Retaux Citation2008). Although many lamprey species actively feed on other fishes as juveniles before maturation, the larva of non-parasitic species transform directly into non-feeding adults, spawn, and die. This parasitic/non-parasitic dichotomy is reflected in the taxonomic diversity of lampreys, in which most parasitic species are paired with a non-parasitic derivative (Pereira et al. Citation2010). Although such ‘satellite’ species occur in most genera of lampreys, the three species pairs of the genus Ichthyomyzon exquisitely illustrate this pattern.
The southern brook lamprey (Ichthyomyzon gagei) is a non-parasitic species that is considered to be a derivative of the parasitic chestnut lamprey (I. castaneus), with which it co-occurs in the southeast United States (Warren & Burr Citation2014). The life history of I. gagei has been summarized, its karyotype has been described (Howell & Duckett Citation1971), and its phylogenetic relationships have been inferred from mitochondrial cytochrome b sequences (Lang et al. 2009).
We report herein the mitochondrial genome of Ichthyomyzon gagei and its phylogenetic relationship as inferred from the entire mitogenome sequence. The specimen was an adult male collected from Cadron Creek, Arkansas, (35.1936N; 92.5302W), ethanol fixed, and vouchered at the University of Southern Indiana Natural History Collection (# R2146). DNA was extracted from muscle tissue following conventional proteinase K and phenol–chloroform extraction protocols. Mitochondrial DNA was amplified by primer-walking and the individual PCR fragments were sequenced.
The mitogenome of Ichthyomyzon gagei (Genbank: KY056640) is 16,359 bp in length and is slightly longer than those reported for I. fossor (16,150 bp) and I. unicuspis (16,163 bp). Most of the length differences are due to variation in the non-coding regions (NC1 and NC2), although occasional insertions/deletions were found in tRNA and rRNA genes. Base composition (L-strand) was similar to that previously reported for other vertebrates with a low guanine content (12.8%) and almost equal frequencies of adenine (33.1%), cytosine (23.8%) and thymine (30.3%) residues. The mitogenome has the same genic arrangement as Petromyzon marinus (Lee & Kocher Citation1995) and includes 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs and 2 non-coding regions. The percent identities of the protein-coding genes from I. gagei and the two other available species of Ichthyomyzon ranged from 90% (ND4) to 94% (CO1, CO2, ATP8, CO3, and ND3). Comparison with P. marinus revealed greater divergences (81% ND3 to 89% CO2 and CO3).
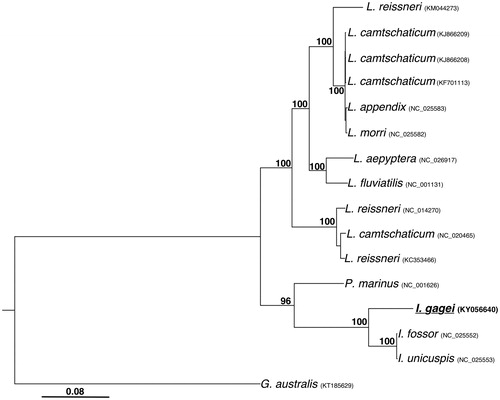
Phylogenetic analyses of the 16 available lamprey mitogenomes were consistent with previous relationships inferred from mitogenomic data (Lang et al. 2009; Ren et al. Citation2015; ). Ichthyomyzon gagei is sister to I. fossor and I. unicuspis, while the three available species of the genus Ichthyomyzon form the sister group to the monotypic genus Petromyzon. The remainder of petromyzontid lampreys form a well-supported group that is sister to the Petromyzon + Ichthyomyzon clade.
Figure 1. Phylogenetic analysis of lamprey mitogenomes. Full length mitogenome sequences were aligned using MUSCLE (Edgar Citation2004). Tree was inferred from both maximum parsimony (PAUP*; Swofford Citation2002) and maximum likelihood analysis with a HKY85 substitution model (PhyML; Guindon et al. Citation2010). Bootstrap values (1000 replicates) are indicated at each node. The current species under investigation (I. gagei, Genbank: KY056640) is underlined and bold. GenBank accession numbers are included for each sequence.
Disclosure statement
The authors declare no conflict of interests. This research was funded by institutional support from the University of Southern Indiana (KJD).
References
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Howell WM, Duckett CR. 1971. Somatic chromosomes of the lamprey, Ichthyomyzon gagei (Agnatha: Petromyzontidae). Experientia. 27:222–223.
- Lang NJ, Roe KJ, Renaud CB, Gill HS, Potter IC, Freyhof J, Naseka AM, Cochran P, Perez HE, Habit EM, et al. 2009. Novel relationships among lampreys (Petromyzontiformes) revealed by a taxonomically comprehensive molecular data set. American Fisheries Society Symposium. 72:41–55.
- Lee WJ, Kocher TD. 1995. Complete sequence of a sea lamprey (Petromyzon marinus) mitochondrial genome: early establishment of the vertebrate genome organization. Genetics. 139:873–887.
- Osorio J, Retaux S. 2008. The lamprey in evolutionary studies. Dev Genes Evol. 218:221–235.
- Pereira AM, Robalo JI, Freyhof J, Maia C, Fonseca JP, Valente A, Almada VC. 2010. Phylogeographical analysis reveals multiple conservation units in brook lampreys Lampetra planeri of Portuguese streams. J Fish Biol. 77:361–371.
- Ren J, Pu J, Buchinger T, Zhu X, Baker C, Li W. 2015. The mitogenomes of the pouched lamprey (Geotria australis) and least brook lamprey (Lampetra aepyptera) with phylogenetic considerations. Mitochondrial DNA a DNA Mapp Seq Anal. 27:3560–3562.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland (MA): Sinauer Associates.
- Warren ML, Burr BM. 2014. Freshwater fishes of North America: Petromyzontidae to Catostomidae. Baltimore (MD): Johns Hopkins University Press.
