Abstract
The complete mitochondrial genome of Ellobium chinense (Ellobioidea, Ellobiidae), an Endangered species in South Korea, is reported here for the first time. The mitogenome of E. chinense is 13,979 base pairs in total length and includes 13 PCGs, small and large rRNAs, and 21 tRNAs. Twelve genes are encoded on the light-strand and 24 genes on the heavy-strand. Compared to four other ellobiid species, the PCGs of E. chinense have a conserved gene order except for the positions of ND4L and ND4. These data provide useful molecular information for phylogenetic studies concerning ellobiids and related species.
Ellobium chinense, a species of terrestrial pulmonate gastropod, is designated as an Endangered species in South Korea and is found mainly in the upper intertidal zone (The National Institute of Biological Resources Citation2014). Data are lacking on its biological characteristics and genetic information. In this study, we determined the complete mitochondrial genome of E. chinense (13,979 bp; GenBank accession number KY056647).
A specimen of E. chinense was collected from the upper intertidal zone in Gwangyang-si, Jellanam-do, South Korea. Genomic DNA was extracted (deposited in National Institute of Biological Resources, accession 120913G2) using a QIAmp DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). The mitochondrial genome sequence was obtained using next-generation sequencing, with raw reads generated from an Illumina Hi-seq 2500 (Illumina Inc., San Diego, CA). Using CLC genomic workbench 9.0.1 (CLC Bio-Qiagen, Aarhus, Denmark), 6,282 out of 12,038,798 reads were assembled to produce a circular form of the complete mitochondrial genome with an average of 67 × coverage.
The mitochondrial genome contains large and small rRNAs, 21 tRNAs, and 13 PCGs. tRNAs and PCGs were predicted using tRNAscan-SE 1.21 (Schattner et al. Citation2005), ARWEN (Laslett & Canbäck Citation2008), and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/). The overall base composition of the mtDNA is 24.4% A, 18.6% C, 33.9% T, and 23.1% G. Most of the genes are encoded on the heavy strand, except for four PCGs (ATP6, ATP8, ND3, and COIII) and seven tRNA genes (tRNAGln, tRNALeu, tRNAAsn, tRNAMet, tRNAThr, tRNAGlu, and tRNAArg).
Five protein-coding genes (ND1, ND4, ND4L, ATP8, and COIII) are initiated by the ATG start codon, and six PCGs (COI, COII, ND5, ATP6, CYTB, and ND3) are initiated by TTG. The ND6 and ND2 genes have ATT and GTG as the start codon, respectively. Four genes (ND2, ND3, ND5, and COI) end on the canonical stop codon TAG and another four genes (ND1, ND4, ND6, and COII) use TAA as the stop codon. The remaining five genes (ND4L, CYTB, ATP6, ATP8, and COIII) have an incomplete stop codon and have a terminal T or TA. The tRNAs are spread over the whole genome and their lengths vary from 60 bp to 79 bp. While four other species (Auriculinella bidentata, Ovatella vulcani, Myosotella myositis, and Pedis pedis) in the same family have two tRNAser genes, only one tRNAser has been found on the mitochondrial genome of E. chinense (White et al. Citation2011). The gene order on the E. chinense mitochondrial genome is the same as previously described for two other ellobiid species, A. bidentata and O. vulcani, except for the tRNAser number. In addition, the PCG arrangement of E. chinense is identical to the mitochondrial genome of M. myosotis, except for the position of ND4L, and is identical to P. pedis, except for the position of ND4 (White et al. Citation2011).
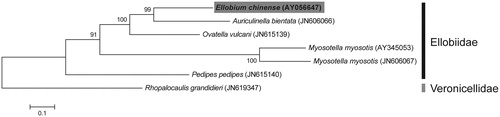
The complete mitochondrial genomes of E. chinense and four other species from the same family were used to reconstruct a maximum-likelihood tree with 500 bootstrap replicates (). The phylogenetic analyses were performed using MEGA 6.0 (Tamura et al. Citation2013).
Figure 1. The molecular phylogenetic tree of E. chinense and other related species in Ellobiidae based on mitochondrial 13 protein-coding gene sequences. The complete mitochondrial genomes were obtained from GenBank and the accession numbers of the sequences are indicated in parentheses after scientific name of each species. The phylogenetic tree constructed by a maximum-likelihood method with 500 bootstrap replicates.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this report.
Additional information
Funding
References
- Laslett D, Canbäck B. 2008. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30: 2725–2729.
- The National Institute of Biological Resources. 2014. Overview of the Endangered wild species. Incheon, South Korea: NIBR.
- White TR, Conrad MM, Tseng R, Balayan S, Golding R, Martins AM, Dayrat BA. 2011. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetics relationships. BMC Evol Biol. 11:295.
