Abstract
Watermelon is one of the worldwide popular summer fruits and well-known for colourful, sweet, and juicy flesh. The complete nucleotide sequence of cultivated watermelon (Citrullus lanatus L. subsp. vulgaris) chloroplast genome has been determined in this study. The genome was composed of 156,906 bp containing a pair of inverted repeats (IRs) of 26,082 bp, which was separated by a large single-copy region of 70,063 bp and a small single-copy region of 17,895 bp. A total of 114 genes were predicted included 80 protein-coding genes, four rRNA genes, and 30 tRNA genes. Phylogenetic analysis revealed that C. lanatus and other three species belonging to the genus Cucumis were closely clustered into a clade, the family Cucurbitaceae.
Watermelon is one of the worldwide popular summer fruits belonging to the family Cucurbitaceae. Its fruits are well-known for colourful, sweet, and juicy flesh. Chloroplasts are organelles mainly present in leaf cell or other green tissue of plant. They contain their own genetic system as well as the entire machinery necessary for the process of photosynthesis. Meanwhile, studies have reported that the watermelon chloroplast genome was associated with the sexual differentiation (Levi et al. Citation2006), and was important for plant taxonomy (Dane et al. Citation2004; Hu et al. Citation2011) and phylogenetic system researches (Dane & Lang Citation2004; Levi & Thomas Citation2005; Dane & Liu Citation2007; Dane et al. Citation2007) in watermelon.
To date, complete chloroplast genome sequences of many species in the family Cucurbitaceae have been published at NCBI Organelles Genome Database, including cultivated cucumber (Kim et al. Citation2006; Chung et al. Citation2007; Pląder et al. Citation2007), wild cucumber (Wu et al. Citation2014), melon (Rodriguez-Moreno et al. Citation2011). However, complete chloroplast sequence of cultivated watermelon has not yet been reported and only several partial chloroplast genome sequences were reported. In this study, we reported complete sequence of chloroplast genome of C. lanatus to provide basic genetic information and help understand genomic diversity of watermelon species.
The genomic DNA was extracted from young leaves of C. lanatus collected from and cultivated in the farm field of Northeast Agricultural University (126°43′16.7′′E, 45°44′23.8′′N), Harbin, China. A pair-end (PE) library with 300 bp insert was constructed and sequenced using an Illumina Hiseq 2000 platform by BGI, Shenzhen, China. Whole genome sequence data of 20 Gb were generated and trimmed, high quality PE reads of 0.4 Gb were randomly extracted and assembled with the BAC end sequences (Guo et al. Citation2013) using SPAdes (v3.6.2) (Bankevich et al. Citation2012), as described in Rodriguez-Moreno et al.(Rodriguez-Moreno et al. Citation2011). Contigs representing the chloroplast genome were retrieved, ordered and joined into a single draft sequence by comparison with the chloroplast genome of Cucumis sativus (GenBank accession no. NC_007144.1) as a reference (Kim et al. Citation2006). The draft sequence was confirmed and manually corrected by PE read mapping. The draft sequence was annotated using an integrated web server, CpGAVAS (Chang et al. Citation2012), and manual curation based on BLAST searches.
The complete chloroplast genome of C. lanatus was double-stranded circular DNA with 156,906 bp in length (GenBank accession no. KY014105). Its structure was similar to most of the described chloroplast genomes from cucurbit plants (Kim et al. Citation2006; Chung et al. Citation2007; Pląder et al. Citation2007; Rodriguez-Moreno et al. Citation2011), composed of two inverted repeated regions (IRa and IRb) of 25,150 bp, which was divided by a large single-copy (SSC) region of 70,063 bp and a small single-copy (LSC) region of 17,895 bp. The genome was typical of AT-rich, and its GC contents were 37.2%. A total of 114 genes were predicted, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes of which 17 genes were duplicated in IR regions.
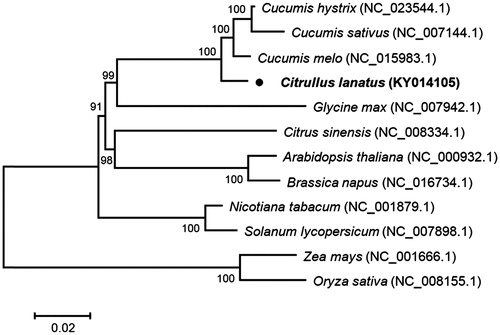
Phylogenetic analysis was conducted with common 53 chloroplast protein-coding sequences of C. lanatus and other 11 species, using a Neighbour-Joining method of MEGA 7.0 with 1000 bootstrap replicates (Tamura et al. Citation2013). The phylogenetic tree indicated that C. lanatus and other three species belonging to the genus Cucumis were closely clustered into a clade, the family Cucurbitaceae, and other species belonging to same family were also well clustered into their corresponding clades, as expected ().
Figure 1. Phylogenetic tree showing relationship between C. lanatus and 11 species belonging to different families. Phylogenetic tree was constructed based on 53 protein-coding genes of chloroplast genomes using Neighbour-Joining method with 1000 bootstrap replicates. Numbers in each of the node indicated the bootstrap support values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chang L, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:1–7.
- Chung SM, Gordon VS, Staub JE. 2007. Sequencing cucumber (Cucumis sativus L.) chloroplast genomes identifies differences between chilling-tolerant and -susceptible cucumber lines. Genome. 50:215–225.
- Dane F, Lang P. 2004. Sequence variation at cpDNA regions of watermelon and related wild species: implications for the evolution of Citrullus haplotypes. Am J Bot. 91:1922–1929.
- Dane F, Lang P, Bakhtiyarova R. 2004. Comparative analysis of chloroplast DNA variability in wild and cultivated Citrullus species. Theor Appl Genet. 108:958–966.
- Dane F, Liu J. 2007. Diversity and origin of cultivated and citron type watermelon (Citrullus lanatus). Genet Resour Crop Ev. 54:1255–1265.
- Dane F, Liu J, Zhang C. 2007. Phylogeography of the bitter apple, Citrullus colocynthis. Genet Resour Crop Ev. 54:327–336.
- Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Zheng Y, Mao L, Ren Y, Wang Z, et al. 2013. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 45:51–58.
- Hu JB, Li JW, Li Q, Ma SW, Wang JM. 2011. The use of chloroplast microsatellite markers for assessing cytoplasmic variation in a watermelon germplasm collection. Mol Biol Rep. 38:4985–4990.
- Kim JS, Jung JD, Lee JA, Park HW, Oh KH, Jeong WJ, Choi DW, Liu JR, Cho KY. 2006. Complete sequence and organization of the cucumber (Cucumis sativus L. cv. Baekmibaekdadagi) chloroplast genome. Plant Cell Rep. 25:334–340.
- Levi A, Thies JA, Ling KS, Harrison HF, Thomas CE, Simmons AM, Hassell R, Keinath AP. 2006. Novel watermelon breeding lines containing chloroplast and mitochondrial genomes derived from the desert species Citrullus colocynthis. Hortscience. 41:463–464.
- Levi A, Thomas CE. 2005. Polymorphisms among chloroplast and mitochondrial genomes of Citrullus species and subspecies. Genet Resour Crop Ev. 52:609–617.
- Pląder W, Yukawa Y, Sugiura M, Malepszy S. 2007. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: its composition and comparative analysis. Cell Mol Biol Lett. 12:584–594.
- Rodriguez-Moreno L, Gonzalez VM, Benjak A, Marti MC, Puigdomenech P, Aranda MA, Garcia-Mas J. 2011. Determination of the melon chloroplast and mitochondrial genome sequences reveals that the largest reported mitochondrial genome in plants contains a significant amount of DNA having a nuclear origin. BMC Genomics. 12:424.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wu Z, Jia L, Shen J, Jiang B, Qian C, Lou Q, Li J, Chen J. 2014. The complete chloroplast genome sequence of the wild cucumber Cucumis hystrix Chakr. (Cucumis, cucurbitaceae). Mitochondrial DNA. 27:1–3.
