Abstract
The complete mitochondrial genome of gilbert’s irish lord (Hemilepidotus gilberti), a fish belonging to family Cottidae, was sequenced for the first time. This complete mitochondrial genome was 16,907 nucleotides in length, consisting of 38 mitochondrial genes (13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region). The order of these genes was similar to that of other teleosts. The overall A, C, G, and T nucleotide contents in mitogenome were 26.8%, 30.4%, 17.0%, and 25.8%, respectively. The A + T content (52.6%) was higher than the G + C content (47.4%). NJ phylogenetic analysis was performed for 10 related species within the family of Cottidae along with, two fish species belonging to another family (Sebastidae).
Genus Hemilepidotus contains six species in the world (Froese & Pauly Citation2016). Among them, Hemilepidotus gilberti belongs to the order of Scorpaeniformes and, family Cottidae (Nakabo Citation2013). Although many marine sculpins are known to copulate, interestingly, this species is a non-copulating sculpin (Hayakawa & Munehara Citation1996). They are distributed from North Pacific including eastern sea of Korea, and Hokkaido, Japan to western Bering Sea (Kim et al. Citation2005; Masuda et al. Citation1984).
In this study, a H. gilberti specimen was collected from a commercial fish market near the East Sea of Korea for complete mitogenome sequencing. The specimen was deposited at the National Marine Biodiversity Institute of Korea (Voucher No. MABIK PI00039348). We dissected the right dorso-lateral muscle of the specimen and preserved it in 95% ethanol. Genomic DNA was extracted from the muscle using Qiagen DNeasy Blood and Tissue kit (Qiagen Korea Ltd, Seoul, South Korea) following the manufacturer’s instructions. The complete mitochondrial DNA was sequenced on the Hiseq2000 platform using next generation sequencing technique (Illumina, San Diego, CA). Geneious 9.1.2 (Biomatters Ltd, Auckland, New Zealand), tRNA Scan-SE1.21 software (http://lowelab.ucsc.edu/tRNAScan-SE/), and MitoFish (Mitochondrial Genome Database of Fish, http://mitofish.aori.u-tokyo.ac.jp/) were used to assemble and annotate the mitochondrial DNA sequences.
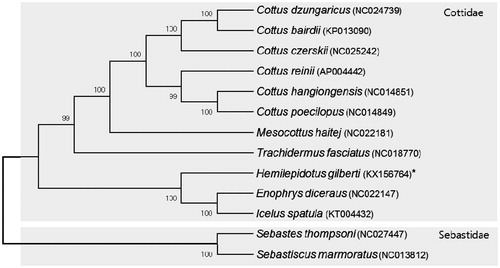
The mitochondrial genome (GenBank Accession No. KX156764) was 16,907 base pairs, in length. A, C, G and T contents were 26.8%, 30.4%, 17.0% and 25.8% respectively. A–T content (52.6%) was slightly higher than G–C content (47.4%). This genome contained 13 protein-coding genes, 2 rRNA genes (12s and 16s RNA), 22 tRNA genes and 1 D-loop region. H. gilberti mitogenome had two rRNA subunits (12S and 16S). They were located between tRNA-Phe and tRNA-Leu (UUR) and separated by tRNA-Val gene. Among tRNAs, two forms of tRNA-Leu (UUR and CUN) and tRNA-Ser (UCN-AGN) were identified. Most genes of H. gilberti mitogenome were encoded on the L-strand except 16S rRNA, ND6 and tRNA (Gln, Ala, Asn, Cys, Tyr, Ser (UCN), Glu and Pro) genes which were encoded on the H-strand. All tRNAs except tRNA-Ser (AGN) have the typical clover leaf structure. The tRNA-Ser (AGN) had a reduced DHU arm. In order to discuss the phylogenetic relationships, the complete mitochondrial genome sequences of 13 species available in the public databases were used to construct a neighbor-joining (NJ) tree using the Kimura two-parameter model (Kimura Citation1980). Its confidence was assessed via 5000 bootstrap replications using Mega 6 (Tamura et al. Citation2013). The construction of phylogenetic tree showed that H. gilbert was clustered in the family of Cottidae and has closer relationship to the Enophrys diceraus and Icelus spatula. They were well distinguished from Family Sebastidae ().
Figure 1. A NJ tree using the coding genes of complete mitochondrial genomes of H. gilberti with another 10 species belonging to family Cottidae and 2 species belonging to family Sebastidae. The complete mitogenome was downloaded from GenBank (accession number indicated after the scientific name of each species). The phylogenetic tree was constructed with MEGA6 using 5000 bootstrap replicates.
Disclosure statement
The authors report no conflicts of interest. The authors are solely responsible for the content and the writing of the paper.
Additional information
Funding
References
- Froese R, Pauly D., editors. 2016. FishBase, version (06/2016) [Internet]. Available from: www.fishbase.org.
- Hayakawa Y, Munehara H. 1996. Non-copulatory spawning and female participation during early egg care in a marine sculpin Hemilepidotus gilberti. Ichthyol Res. 43:73–78.
- Kim IS, Choi Y, Lee YJ, Kim BJ, Kim JH. 2005. Illustrated book of Korean fishes. Seoul, Korea: Kyo-Hak Publishing. p. 613. (In Korean).
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120.
- Masuda H, Amaoka K, Araga C, Uyeno T, Yoshino T. (1984). The fishes of the Japanese Archipelago, vol. 1. Tokyo, Japan: Tokai Univ Press. p. 437.
- Nakabo T., editor. 2013. Family Cottidae. In: Hatooka K., editor. Fishes of Japan with pictorial keys to the species, 3rd ed. Hadano: Tokai Univ Press. p. 1160–1188. (In Japanese).
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729.
