Abstract
Dipterocarpaceae are one of the economically most important native tree families for timber production in tropical Asia. We report the complete chloroplast genome of Vatica odorata (Griff.) Symington, the first in the family Dipterocarpaceae. The chloroplast genome was 151,465 bp in length, with a large single-copy (LSC) region of 83,538 bp and a small single-copy (SSC) region of 20,095 bp, separated by two inverted repeat (IRs) regions of 23,916 bp. It contained 126 genes, including 90 coding genes, 30 tRNA genes, and 8 rRNA genes. The overall GC content was 37.2%, and 43.1%, 35.2%, and 33.3% in the IRs, LSC and SSC regions, respectively. A phylogenetic tree showed Vatica accumulated more variation when compared with Tilia, and that internal relationships in Malvales need to be reassessed.
Dipterocarpaceae, comprising more than 550 species, are spread throughout tropical Asia, Africa, and South America (Gottwald & Parameswaran Citation1966; Ashton Citation1982), with their centre of diversity in Malaysia (Ashton Citation1988). In China, Dipterocarpaceae are represented by 5 genera and 12 species (Xi-wen et al. Citation2007).
Despite a number of previous studies using single or multiple marker approaches (both cp- and nDNA) for Dipterocarpaceae (Kajita et al. Citation1998; Gamage et al. Citation2006; Yulita Citation2013), no studies to date have generated and used complete genomes (nuclear or chloroplast).
The genus Vatica comprises about 65 species (Xi-wen et al. Citation2007). Its timber is especially valuable as it is very hard and resistant to insect attacks. However, at the current rate of forest exploitation, 65 million ha of forest are projected to be cleared for agriculture between 2012 and 2030 (FAO Citation2012). Here, we report the complete chloroplast sequence of Vatica odorata to provide genomic resources for timber identification and quality improvement.
Genomic DNA of one individual of Vatica odorata was extracted from fresh leaves collected in XTBG Botanical Garden (22°35′24″N, 99°30′01″E Yunnan, China – voucher STRIJK_1594 deposited in the herbarium of the College of Forestry of Guangxi University, Nanning, China), using a SDS protocol (modified from Healey et al. Citation2014). Library construction and sequencing were performed by Novogene (Beijing, China), following Illumina HiSeq2500 system manufacturer instructions. De novo assembly of the cp genome was conducted using org.asm v0.2.05 (ORG.ASM Citation2016) and annotation using cpGAVAS (Liu et al. Citation2016).
The complete cp genome of Vatica odorata (GenBank accession KX966283) was 151,465 bp in length, with a large single-copy (LSC) region of 83,538 bp and a small single-copy (SSC) region of 20,095 bp, separated by two inverted repeat (IRs) regions of 23,916 bp. The genome contained 126 genes, including 90 coding genes, 30 tRNA genes, and 8 rRNA genes. The overall GC content of the cp genome was 37.2% and 43.1% in IRs, which was greater than LSC (35.2%) and SSC (33.3%) regions.
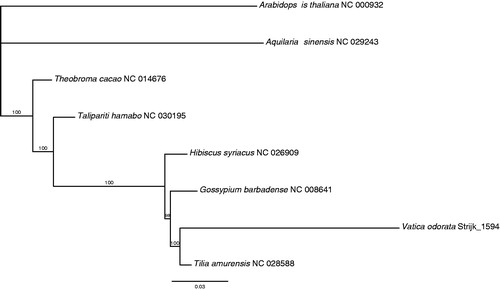
The chloroplast genome of Vatica odorata is the first to be published for the family and to confirm the phylogenetic position within the broader Malvales order, we reconstructed phylogenetic relationships using other plastome sequences available in GenBank using PHYML (Guindon et al. Citation2005). Phylogenetic tree shows the particularly long branch of Vatica, as opposed to Tilia, and the short branches underlying the nodes of the preceding clades (). However, Malvaceae in its current circumscription are not monophyletic. Our results also suggest that the merger of Tiliaceae with other Malvaceae (sensu Chase & Reveal Citation2009) does not aid in resolving the phylogenetic quagmire in the family.
Figure 1. ML phylogenetic tree of the six Malvales in available chloroplast sequences in GenBank, plus the chloroplast sequence of Vatica odorata. The tree is rooted with the Brassicales (Arabidopsis thaliana). Bootstraps (100 replicates) are shown at the nodes. Scale in substitution per site.
This is the first study to report the complete chloroplast genome of a Dipterocarpaceae species and will be of great interest in further studies on genomic diversity, origin, and evolution of Dipterocarpaceae.
Acknowledgements
We would like to acknowledge B. Kingsley, M. Roeder and the horticultural staff of Xishuangbanna Tropical Botanical Garden (Chinese Academy of Sciences) for their assistance in sampling material of Vatica odorata.
Disclosure statement
This work was supported by grants from Guangxi University (Nanning), the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (GXU) and the provincial government of Guangxi Province (“100 Talents” Program; recruitment of overseas talents for colleges and universities in Guangxi) to JSS, and grants from China Postdoctoral Science Foundation (No. 2015M582481 and 2016T90822) to DDH. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Ashton PS. 1982. Dipterocarpaceae. Flora malesiana. Series 1, Vol. 9. The Hague: Martinus-Nijhoff Publishers.
- Ashton PS. 1988. Dipterocarp biology as a window to the understanding of tropical forest structure. Annu Rev Ecol Syst. 19:347–370.
- Chase MW, Reveal JL. 2009. A phylogenetic classification of the land plants to accompany APG III. Bot J Linn Soc. 161:122–127.
- FAO. 2012. Supporting sustainable forest management through the Global Forest Resources Assessments Long-Term Strategy (2012–2030). Rome: FAO.
- Gamage DT, Silva MPd, Inomata N, Yamazaki T, Szmidt AE. 2006. Comprehensive molecular phylogeny of the subfamily Dipterocarpoideae (Dipterocarpaceae) based on chloroplast DNA sequences. Genes Genet Syst. 81:1–12.
- Gottwald H, Parameswaran N. 1966. Das sekundare xylem der familie dipterocarpaceae, anatomische untersuchungen zur taxonomie und phylogenie. Bot Jahrb Syst. 85:410–508.
- Guindon S, Lethiec F, Duroux P, Gascuel O. 2005. PHYML Online – a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559.
- Healey A, Furtado A, Cooper T, Henry RJ. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 10:1.
- Kajita T, Kamiya K, Nakamura K, Tachida H, Wickneswari R, Tsumura Y, Yoshimaru H, Yamazaki T. 1998. Molecular phylogeny of Dipterocarpaceae in Southeast Asia based on nucleotide sequences of matK, trnL intron, and trnL-trnF intergenic spacer region in chloroplast DNA. Mol Phyl Evol. 10:202–209.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2016. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- ORG.ASM. 2016. ORG.ASM: organellar assembler [Internet]; [cited 2016 Nov 1]. Available from: http://pythonhosted.org/ORG.asm/
- Xi-wen L, Jie L, Ashton PS. 2007. Flora of China: Dipterocarpaceae. vol 13. Missouri/Beijing: Missouri Botanical Garden Press & Science Press Beijing.
- Yulita KS. 2013. Secondary structures of chloroplast trnL intron in Dipterocarpaceae and its implication for the phylogenetic reconstruction. HAYATI J Biosci. 20:31–39.
