Abstract
Magnoliaceae are both economically and ornamentally important trees. Despite extensive studies in this family, the taxonomy of Michelia L. remains unclear, as well as the taxonomical status of Michelia alba. Herein, we report the complete chloroplast genome of M. alba DC. The chloroplast genome was 159,789 bp in length, with a large single-copy (LSC) region of 87,951 bp and a small single-copy (SSC) region of 18,798 bp, separated by two inverted repeat (IRs) regions of 26,570 bp. It contained 156 genes, including 83 coding genes, 68 tRNA genes, and 8 rRNA genes. The overall GC content was 39.3%, and 43.2%, 38.0%, 34.3%, in the IRs, LSC, and SSC regions, respectively. A phylogenetic analysis showed that M. alba is closely related to M. odora, with the genus Michelia nested inside Magnolia.
The Magnoliaceae comprise ca. 220–240 species (Nie et al. Citation2008), widely used for ornamental urban planting all around the world, especially the tropical and subtropical species for their fragrant flowers. Michelia L. (≈30 species) is similar to Magnolia L., except that flowers are clustered among leaves and not terminal as in Magnolia, and recent molecular data suggested that the former could be a synonym of the later (Nie et al. Citation2008). Michelia alba D.C. is an evergreen tall tree native to Indonesia and now widespread in tropical and subtropical countries (Nooteboom Citation1985) for it fragrant flowers. Because wild or fruiting trees are unknown, M. alba is commonly considered as a hybrid between M. champaca and M. montana (Nooteboom Citation1985). However, no molecular study has investigated the taxonomical status of M. alba (but see Nie et al. Citation2008, where M. alba was closely related to M. champaca). Here, we report the complete chloroplast sequence of M. alba to provide resources for delineation of the taxonomical status of this species and its potential horticultural varieties.
We extracted genomic DNA from fresh M. alba leaves collected from the Guangxi University (GXU) campus (22°50'20.5”N, 108°17'23.1”E Nanning, China – voucher HINSINGER_2016-NNG1 and deposited in the herbarium of the College of Forestry of Guangxi University, Nanning, China), using a modified SDS protocol (Hinsinger & Strijk Citation2015; Cvetkovic et al. Citation2016). Library construction and sequencing were performed by Novogene (Beijing, China), according to the Illumina HiSeq2500 system manufacturer instructions. We performed a de novo assembly of the chloroplast genome with org.asm v0.2.05 (http://pythonhosted.org/ORG.asm/) followed by manual adjustments and a iterative mapping step (see Hinsinger and Strijk, Citation2016) to correct for potential assembly mistakes. We used Geneious R9 v9.1.6 (Biomatters Ltd, Auckland, New Zealand) to transfer the annotations from the congeneric M. odora sequence (NC023239).
The complete cp genome of M. alba (GenBank accession KY204085) was 159,789 bp in length, with a 87,951 bp large single-copy (LSC) region and a 18,798 bp small single-copy (SSC) region, separated by 2 26,570 bp inverted repeat (IRs). It contained 156 genes, including 83 CDS, 68 tRNA genes, and 8 rRNA genes. The overall GC content was 39.3%, and 43.2%, 38.0%, 34.3% in the IRs, LSC, and SSC, respectively.
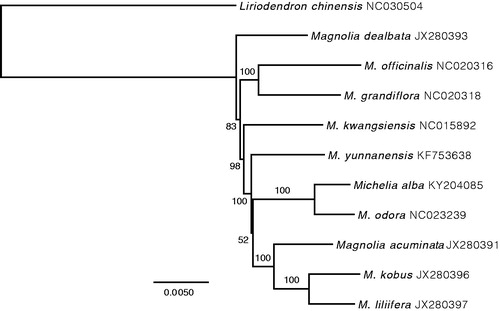
We used RaxML as implemented in Geneious R9 v9.1.6 to construct a Maximum-Likelihood phylogenetic tree including 10 available representative species in Magnoliaceae (). As expected, M. alba was closely related to M. odora, forming a clade included in Magnolia. Bootstrap values were high for all nodes but the relationship between M. yunnanensis and the ((M. kobus, M. liliifera), M. acuminata) clade. Interestingly, this grouping was already poorly supported in Nie et al. (Citation2008), suggesting the lack of informative sites was not the cause of this low value. Finally, the placement of M. liliifera as the sister taxa of M. kobus suggested a mistaken labelling for M. liliiflora, as this species was closely related to M. kobus in Nie et al. (Citation2008).
Figure 1. ML phylogenetic tree of 10 selected Magnoliaceae chloroplast sequences, plus the chloroplast sequence of M. alba. The tree is rooted with Liriodendron chinensis. Bootstraps (1000 replicates) are shown at the nodes. Scale in substitution per site.
We expect this sequence will help to clarify the hybrid status of M. alba, and provide additional genomic resources for Magnoliaceae studies.
Acknowledgements
We would like to acknowledge the horticultural staff of the Guangxi University (GXU) for their assistance in sampling material of M. alba.
Disclosure statement
This work was supported by grants from Guangxi University (Nanning), the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (GXU) and the provincial government of Guangxi Province (‘100 Talents’ Program; recruitment of overseas talents for colleges and universities in Guangxi) to JSS, and grants from China Postdoctoral Science Foundation (No. 2015M582481 and 2016T90822) to DDH. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cvetkovic T, Hinsinger DD, Strijk JS. 2016. The first complete chloroplast sequence of a major tropical timber tree in the Meranti family: Vatica odorata (Dipterocarpaceae). Mitochondrial DNA. in press. doi: 10.1080/23802359.2016.1275837.
- Hinsinger DD, Strijk JS. 2015. Complete chloroplast genome sequence of Castanopsis concinna (Fagaceae), a threatened species from Hong Kong and South-Eastern China. Mitochondrial DNA. 1–2. doi: 10.3109/19401736.2015.1110800.
- Hinsinger DD, Strijk JS. 2016. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. doi: 10.1016/j.plgene.2016.08.002.
- Nie ZL, Wen J, Azuma H, Qiu YL, Sun H, Meng Y, Sun WB, Zimmer EA. 2008. Phylogenetic and biogeographic complexity of Magnoliaceae in the Northern Hemisphere inferred from three nuclear data sets. Mol Phylogenet Evol. 48:1027–1040.
- Nooteboom HP. 1985. Notes on Magnoliaceae, with a revision of Pachylarnax and Elmerrillia and the Malesian species of Manglietia and Michelia. Blumea. 31:65–121.
