Abstract
In this study, the complete mitochondrial genome of the White-Nose Syndrome pathogen, Pseudogymnoascus destructans (=Geomyces destructans), is sequenced. The circular mitochondrial genome is 32,181 bp long and encodes 13 standard proteins, 2 ribosomal RNA subunits, and 27 tRNAs. The genome contains two introns located within the cytochrome c oxidase subunit 1 gene (cox1) and the large ribosomal RNA subunit (rrnl), with each intron encoding one gene, orf110 and rps3, respectively. Phylogenetic analysis of the concatenated mitochondrial protein-coding genes of P. destructans and close representatives in Leotiomycetes showed that P. destructans was closely related to Pseudogymnoascus pannorum, consistent with nuclear genes.
The White-Nose Syndrome (WNS) is a devastating infection of North American bats, caused by the fungal pathogen, Pseudogymnoascus destructans (Minnis & Lindner Citation2013). The infection is characterized by a mycosal infection of epithelial tissues (Meteyer et al. Citation2009), which eventually leads to electrolyte imbalance, evaporative water loss, frequent arousal from hibernation (Willis et al. Citation2011), and starvation (Cryan et al. Citation2010). Since 2006, the WNS epidemic has spread to 29 US States and 5 Canadian Provinces, with over 6 million cases of bat mortality attributed to P. destructans (US Fish and Wildlife Service Citation2016).
The data published on P. destructans genomes by Cuomo et al. (Citation2010) and Drees et al. (Citation2016) contained incompletely assembled and un-annotated mitochondrial DNA sequences. Here, we present the complete mitochondrial genome sequence of P. destructans strain 20631-21, the initial case of WNS was from William’s Hotel Mine in New York, USA (Blehert et al. Citation2009). This sample was collected bat by the US Forestry Service from a deceased little brown in 2008 (ATTC: MYA-4855). We used an Illumina MiSeq platform to sequence the full genome of P. destructans and mitochondrial sequences were extracted from our raw reads through in silico baiting, trimmed for quality using MIRA (V 4.0.2) (Hahn et al. Citation2013), and merged overlapping paired end reads using FLASh (V 1.2.11) (Magoč & Salzberg Citation2011). The complete mitochondrial sequence was assembled using the MITObim pipeline (V 1.8) (Hahn et al. Citation2013), yielding a circular molecule of 32,182 bp, with a GC content of 28.5%. The genome was built based on 46,037 paired-end reads with an average quality of 84 and an average coverage 510×.
The mitochondrial genome was annotated through the MITOS web server, with 16 predicted open reading frames (ORFs) which overlapped with functional proteins (Wheeler et al. Citation2003; Camacho et al. Citation2008; Bernt et al. Citation2013) (Figure S1). The full mitochondrial genome encodes 13 genes of the oxidative phosphorylation system, the small and large ribosomal RNA subunits (rns and rnl respectively), and 27 tRNA genes (GenBank Accession: KY318514.1). A 112 amino acid fragment of the 5′ end of the cox1 gene (labeled as cox1-1) is separated from the rest of cox1 (labeled as cox1-2) by a 1330 bp intron, itself containing the intronic gene orf110, encoding the protein domain for catalytic GIY-YIG and putative intron-encoded endonuclease bI1. An additional intron was found within the large ribosomal RNA subunit, which codes for ribosomal protein S3 (rps3). A 122bp fragment of the atp9 gene was found within P. destructans mitochondria, although this sequence was not found within the P. pannorum mitochondrial genome (Zhang et al. Citation2016).
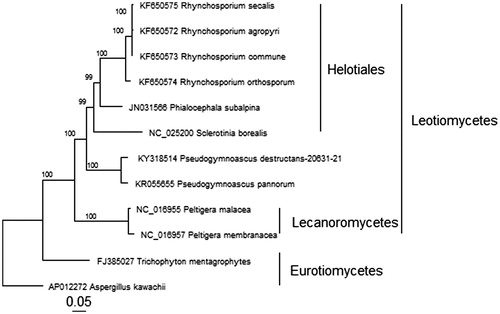
We also present the phylogenetic relationships among 12 representative sequenced filamentous ascomyceteous species (), based on concatenated nucleotide sequences of 13 mitochondrial protein-coding genes aligned using MAFFT (V 7.205) (Katoh & Standley Citation2013). The phylogenetic analysis was completed using a Maximum Likelihood approach with a GTRGAMMA model of nucleotide substitution and rate heterogeneity with RAxML (V 8.0.25) (Stamatakis Citation2014) and visualized using the R package ggtree (Yu et al. Citation2016).
Figure 1. Phylogenetic relationship between P. destructans and representatives of related ascomycete species based on concatenated nucleotide sequences of 13 protein-coding genes: atp6, atp8, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6 for a total of 10,372 characters. All non-P. destructans sequences were obtained from GenBank with accession numbers shown before the species names.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, Coleman JTH, Darling SR, Gargas A, Niver, et al. 2009. Bat white-nose syndrome: an emerging fungal pathogen? Science. 323:227
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2008. BLAST+: architecture and applications. BMC Bioinformatics. 10:421.
- Cryan PM, Meteyer CU, Boyles JG, Blehert DS. 2010. Wing pathology of white-nose syndrome in bats suggests life-threatening disruption of physiology. BMC Biol. 8:135.
- Cuomo CA, Blehert DS, Lorch JM, Young SK, Zeng Q, Pearson M, Alvarado L, Berlin A, Chapman S, Chen Z, et al. 2010. The Genome Sequence of Geomyces destructans strain 20631-21. Unpublished. The Broad Institute Genome Sequencing Platform.
- Drees KP, Palmer JM, Sebra R, Lorch JM, Chen C, Wu CC, Bok W, Keller NP, Blehert DS, Cuomo, et al. 2016. Use of multiple sequencing technologies to produce a high-quality genome of the fungus Pseudogymnoascus destructans, the causative agent of bat white-nose syndrome. Genome Announc. 4:e00445-16.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res, gkt371.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 27:2957–2963.
- Meteyer CU, Buckles EL, Blehert DS, Hicks AC, Green DE, Shearn-Bochsler V, Thomas NJ, Gargas A, Behr MJ. 2009. Histopathologic criteria to confirm white-nose syndrome in bats. J Vet Diagn Invest. 21:411–414.
- Minnis AM, Lindner DL. 2013. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of Eastern North America. Fungal Biol. 117:638–649.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- US Fish and Wildlife Service. 2016. White-Nose Syndrome: The devastating disease of hibernating bats in North America. Available from: www.whitenosesyndrome.org
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, Wagner L. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 31:28–33.
- Willis CK, Menzies AK, Boyles JG, Wojciechowski MS. 2011. Evaporative water loss is a plausible explanation for mortality of bats from white-nose syndrome. Integr Comp Biol. 51:364–373.
- Yu G, Smith D, Zhu H, Guan Y, Lam TTY. 2016. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. doi:10.1111/2041-210X.12628.
- Zhang YJ, Huo LQ, Zhang S, Liu XZ. 2016. The complete mitochondrial genome of the cold-adapted fungus Pseudogymnoascus pannorum syn. Geomyces pannorum. Mitochondrial DNA Part A. 27:2566–2567.
