Abstract
Solanum berthaultii is a wild species belonging to Solanaceae family. The complete chloroplast genome of S. berthaultii was constituted by de novo assembly using a small amount of whole genome sequencing data. The chloroplast genome of S. berthaultii was 155,533 bp in length and consisted of 25,593 bp of a pair of inverted repeats, 18,372 bp of small single copy and 85,975 bp of large single-copy regions. 158 genes were annotated including 105 protein-coding, 45 tRNA, and 8 rRNA genes. Maximum-likelihood phylogenetic analysis with eight Solanaceae species revealed that S. berthaultii is most closely grouped with S. tuberosum.
Solanum berthaultii, a wild diploid species originating from Bolivia, South America is a relative to potato (S. tuberosum). It was identified as a source of resistance to several important pathogens such as Phytophthora infestans, Potato virus Y etc. and used for potato breeding (Ewing et al. Citation2000; Park et al. Citation2009; Rauscher et al. Citation2010; Tan et al. Citation2010; Nouri-Ellouz et al. Citation2016). However, interspecific sexual hybridization is limited due to its sexual incompatibility with S. tuberosum caused by the different ploidy levels of the genome and endosperm balance number (Ortiz & Ehlenfeldt Citation1992; Cho et al. Citation1997). Indeed, crop improvement via protoplast fusion with the two different species has been attempted to overcome sexual barriers for interspecific gene transfer (Bidani et al. Citation2007; Nouri-Ellouz et al. Citation2016) and it is necessary to confirm whether or not plastome fusion occurs with sequence information of chloroplast in potato breeding program (Cho & Park Citation2016; Cho et al. Citation2016).
The S. berthaultii (PI310981) was provided by Highland Agriculture Research Institute, South Korea. An Illumina paired-end (PE) genomic library was constructed with total genomic DNA according to the PE standard protocol (Illumina, San Diego, CA) and sequenced using an Illumina HiSeq2000 at Macrogen (http://www.macrogen.com/kor/). Low-quality bases with raw scores of 20 or less were removed and then approximately 2.5 Gbp of high-quality of PE reads were assembled by a CLC genome assembler (CLC Inc, Rarhus, Denmark) (Kim et al. Citation2015). The principal contigs representing the chloroplast genome were retrieved from the total contigs using Nucmer (Kurtz et al. Citation2004) with the chloroplast genome sequence of S. tuberosum (KM489056) as the reference sequence (Cho et al. Citation2016). The representative chloroplast contigs were arranged in order based on BLASTZ analysis (Schwartz et al. Citation2003) with the reference sequence and connected to a single draft sequence by joining the overlapping terminal sequences. The chloroplast genes were predicted using DOGMA (Wyman et al. Citation2004) and BLAST searches.
The complete chloroplast genome of S. berthaultii (GenBank accession no. KY419708) was 155,533 bp in length including 25,593 bp inverted repeats (IRa and IRb) regions separated by small single copy (SSC) region of 18,372 bp and large single copy (LSC) region of 85,975 bp with the typical quadripartite structure of most plastids, and the structure and gene features were typically identical to those of higher plants. A total of 158 genes with an average size of 583 bp were annotated including 105 protein-coding genes with an average size of 765 bp, 45 tRNA genes, and 8 rRNA genes. An overall GC content was 37.88%.
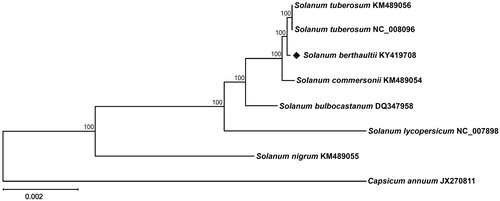
Phylogenetic analysis was performed using chloroplast coding sequences of S. berthaultii and those of seven species including S. lycopersicum (Daniell et al. Citation2006) and Capsicum annuum (Jo et al. Citation2011) in Solanaceae family by a maximum-likelihood method in MEGA 6.0 (Tamura et al. Citation2013). According to the phylogenetic tree, S. berthaultii belonged to the same clade in Solanum species as expected and interestingly it was most closely grouped with S. tuberosum ().
Disclosure statement
The author reports no conflicts of interest. The author alone is responsible for the content and writing of this article.
Additional information
Funding
References
- Bidani A, Nouri-Ellouz O, Lakhoua L, Sihachakr D, Cheniclet C, Mahjoub A, Drira N, Gargouri-Bouzid R. 2007. Interspecific potato somatic hybrids between Solanum berthaultii and Solanum tuberosum L. showed recombinant plastome and improved tolerance to salinity. Plant Cell Tiss Organ Cult. 91:179–189.
- Cho HM, Kim-Lee HY, Om YH, Kim JK. 1997. Influence of endosperm balance number (EBN) in interploidal and interspecific crosses between Solanum tuberosum dihaploids and wild species. Korean J Breed. 29:154–161.
- Cho K-S, Cheon K-S, Hong S-Y, Cho J-H, Im J-S, Mekapogu M, Yu Y-S, Park T-H. 2016. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Rep. 35:2113–2123.
- Cho K-S, Park T-H. 2016. Complete chloroplast genome sequence of Solanum nigrum and development of markers for the discrimination of S. nigrum. Hortic Environ Biotechnol. 57:69–78.
- Daniell H, Lee S-B, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen PK. 2006. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet. 112:1503–1518.
- Ewing EE, Simko I, Smart CD, Bonierbale MW, Mizubuti ESG, May GD, Fry WE. 2000. Genetic mapping from field tests of qualitative and quantitative resistance to Phytophthora infestans in a population derived from Solanum tuberosum and Solanum berthaultii. Mol Breed. 6:25–36.
- Jo YD, Park J, Kim J, Song W, Hur CG, Lee YH, Kang BC. 2011. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 30:217–229.
- Kim K, Lee SC, Lee J, Lee H, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12
- Nouri-Ellouz O, Triki MA, Jbir-Koubaa R, Louhichi A, Charfeddine S, Drira N, Gargouri-Bouzid R. 2016. Somatic hybrids between potato and S. berthaultii show partial resistance to soil-borne fungi and potato virus Y. J Phythpathol. 164:485–496.
- Ortiz R, Ehlenfeldt MK. 1992. The importance of endorsperm balance number in potato breeding and the evolutino of tuber-bearing Solanum species. Euphytica. 60:105–113.
- Park T-H, Foster S, Brigneti G, Jones JDG. 2009. Two distinct potato late blight resistance genes from Solanum berthaultii are located on chromosome 10. Euphytica. 165:269–278.
- Rauscher G, Simko I, Mayton H, Bonierbale M, Smart CD, Grünwald NJ, Greenland A, Fry WE. 2010. Quantitative resistance to late blight from Solanum berthaultii cosegregates with Rpi-ber: insights in stability through isolates and environment. Theor Appl Genet. 121:1553–1567.
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13:103–107.
- Tamura K, STecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tan MYA, Hutten RCB, Visser RGF, van Eck HJ. 2010. The effect of pyramiding Phytophthora infestans resistance genes R Pi-mcd1 and R Pi-ber in potato. Theor Appl Genet. 121:117–125.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.