Abstract
In this study, we determined the mitogenome sequence of Ardeacarus ardeae (Canestrini, 1878) in the family Pterolichidae (Acari, Sarcoptiformes), which is the first complete mitogenome sequence in feather mite. The mitogenome of A. ardeae is 14,069 bp in length and contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a control region (CR). The phylogenetic tree shows that A. ardeae belong to the supercohort Desmonomatides within the order Sarcoptiformes.
Feather mites are referred to as commensal or parasitic species that live on the plumage and skin of birds (Mironov Citation2003; Proctor Citation2003). Ardeacarus ardeae (Canestrini, 1878) (Acari, Acariformes, Sarcoptiformes, Pterolichidae), which is a monotypic species of the genus Ardeacarus Dubinin, 1951, has a worldwide distribution and is mostly observed on wing feathers of the birds in the family Ardeidae (Gaud Citation1981; Gaud & Atyeo Citation1996). Here, we present the complete mitogenome sequence of A. ardeae in family Pterolichidae, which is the first complete mitogenome from a feather mite.
A single specimen of A. ardeae was collected from a wing feathers of little egret Egretta garzetta collected from Eumseong-gun, Chungcheongbuk-do Province (36°56′N, 127°41′E). Mitochondrial DNA extraction, sequencing and gene annotation were performed according to the methods described by Song et al. (Citation2016). The extracted mitochondrial DNA (Inhaevodevo-HYD-0030) was deposited in the Inha University, Incheon, South Korea. The phylogenetic tree was constructed based on the analysis of maximum likelihood (GTR + I + G model, PhyML 3.0) and Bayesian inference (GTR + I + G model, MrBayes 3.2.3) methods using the concatenated sequence of 13 protein-coding genes from the related 15 species within the superfamily Acariformes.
The complete mitogenome of A. ardeae (GenBank accession no. KY352304) was 14,069 bp in length and contained the typical set of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs) and a control region (CR). The overall base composition of the entire A. ardeae mitogenome consisted of 29.3% A, 45.5% T, 10.8% C, 14.5% G and 74.8% AT. Twelve of the PCGs use an ATN codon (N, any nucleotide), while atp8 uses TTG. The stop codons of three PCGs are incomplete and use just one T nucleotide.
The gene arrangement of the A. ardeae mitogenome, with the exception of tRNAs, was found to be almost identical to those of previously published spemicies in Sarcoptiformes. However, A. ardeae has a shorter mitogenome than two closely related species, Dermatophagoides farinae (14,266 bp) and D. pteronyssinus (14,203 bp). These length differences are mainly due to the CR, which is 214 bp long in A. ardeae, while D. farinae and D. pteronyssinus have CRs composed of 410 and 286 bp, respectively (Dermauw et al. Citation2009; Klimov & Oconnor Citation2009).
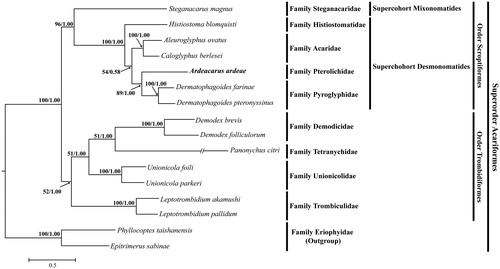
We inferred the phylogenetic relationship of A. ardeae to the Acariformes species. The phylogenetic tree of maximum likelihood and Bayesian inference showed that A. ardeae belongs to the supercohort Desmonomatides within the order Sarcoptiformes. Furthermore, A. ardeae of the family Pterolichidae showed a sister-group relationship with the family Pyroglyphidae (D. farinae and D. pteronyssinus) (). The phylogenetic relationships found in our study are well-congruent with previous mitogenome studies reported by Lee and Wang (Citation2016) and Xue et al. (Citation2016).
Figure 1. Maximum likelihood and Bayesian inference tree based on the mitogenome sequences of Ardeacarus ardeae (KY352304; this study) and 15 other Acariformes species was constructed using PhyML 3.0 and MrBayes 3.2.3, respectively. The following mitogenome were used in this analysis: Aleuroglyphus ovatus (KC700022), Caloglyphus berlesei (KF499016), Demodex brevis (KM114225), D. folliculorum (KM114226), Dermatophagoides pteronyssinus (EU884425), D. farinae (NC013184), Histiostoma blomquisti (KX452726), Leptotrombidium pallidum (AB180098), L. akamushi (AB194045), Panonychus citri (HM189212), Steganacarus magnus (EU935607), Unionicola foili (EU856396) and U. parkeri (HQ386015); outgroup: Epitrimerus sabinae (KR604966) and Phyllocoptes taishanensis (KR604967).
Our study determined the complete mitogenome of A. ardeae in the family Pterolichidae for the first time and will be useful for the detailed study of mitogenome evolution and phylogenetic relationships of the feather mite groups in the order Sarcoptiformes.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Dermauw W, Van Leeuwen T, Vanholme B, Tirry L. 2009. The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods. BMC Genomics. 10:107.
- Gaud J. 1981. Acariens Sarcoptiformes plumicoles des oiseaux Ciconiiformes d'Afrique. 1. Introduction et parasites des Ardeidae. Rev Zool Afr. 95:806–828. French.
- Gaud J, Atyeo WT. 1996. Feather mites of the World (Acarina, Astigmata): the supraspecific taxa. Annal Musée Royale Afrique Entrale Sciences Zoologiques. 277:1–193 (text), 1–436 (illustrations).
- Klimov PB, Oconnor BM. 2009. Improved tRNA prediction in the American house dust mite reveals widespread occurrence of extremely short minimal tRNAs in acariform mites. BMC Genomics. 10:598.
- Lee C-C, Wang J. 2016. The complete mitochondrial genome of Histiostoma blomquisti (Acari: Histiostomatidae). Mitochondrial DNA Part B. 1:671–673.
- Mironov SV. 2003. On some problems in the systematics of feather mites. Acarina. 11:29.
- Proctor HC. 2003. Feather mites (Acari: Astigmata): ecology, behavior, and evolution. Annu Rev Entomol. 48:185–209.
- Song J-H, Kim S, Shin S, Min G-S. 2016. The complete mitochondrial genome of the mysid shrimp, Neomysis japonica (Crustacea, Malacostraca, Mysida). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2781–2782.
- Xue X-F, Guo J-F, Dong Y, Hong X-Y, Shao R. 2016. Mitochondrial genome evolution and tRNA truncation in Acariformes mites: new evidence from eriophyoid mites. Sci Rep. 6:18920.
