Abstract
The present study reports the complete mitochondrial genome of the Suwon tree frog Hyla suweonensis from South Korea. This endangered species is endemic to Korea. The circular mitogenome of H. suweonensis includes 16,895 bp length and contains 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a non-coding region, which is the typical gene arrangement found in the available Hylinae mitogenomes. Phylogenetic analysis of six Hyla spp. mitochondrial genomes revealed that H. suweonensis is closer to H. tsinlingensis.
The Suweon tree frog Hyla suweonensis, family Hylidae, is a tree frog endemic to Korea. The distribution of this species is limited, as it is only found in Incheon, Gyeonggi-, Chungcheong-, and Jeollabuk-do in South Korea, and its typical habitats are rice paddies and wetlands (Yang et al. Citation1997; Lee & Park Citation2016). Since H. suweonensis first description in 1980 (Kuramoto Citation1980), much of its historic habitat disappeared and it now depends on anthropogenically altered habitats (Borzée & Jang Citation2015), and in 2012 it was listed as an endangered species I by the Ministry of Environment of Korea. Chun et al. (Citation2012) examined the genetic diversity of Suweon tree frogs based on two fragments of mitochondrial DNA (cytochrome b and cytochrome oxidase subunit I genes), which revealed a low level of genetic diversity and restricted gene flow. In the present study, the complete mitogenome of the Suweon tree frog was sequenced and compared with available Hyla spp. mitogenomes. In addition, phylogenetic analysis based on these mitogenomes was carried out to evaluate the phylogenetic position of H. suweonensis within the genus.
The H. suweonensis (IN589) used in the present study was collected from Pyeongtaek-si, Gyeonggi-do, South Korea and deposited in the National Institute of Biological Resources (NIBR) at Incheon, South Korea. Total genomic DNA was isolated using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol, and the complete mitogenome sequence was determined using the primer-walking approach. Raw sequences were assembled, edited, and corrected by eye in Geneious 8.1.9 (Kearse et al. Citation2012). Protein-coding genes (PCGs) and ribosomal RNAs (rRNAs) were identified using DOGMA WebServer (Wyman et al. Citation2004) and AIRWIN (Laslett & Canbäck Citation2008).
The complete mitochondrial genome of H. suweonensis was 16,895 bp in length (GenBank accession KY419887), and consisted of 13 typical vertebrate PCGs, 22 transfer RNA genes, 2 rRNA genes (for the small and large subunits, i.e. rrnS and rrnL, respectively), and a putative control (D-loop) region, agreeing with the typical vertebrate gene arrangement (Anderson et al. Citation1981). Most of these genes were encoded on the H-Strand, with the exceptions of the ND6 gene and eight tRNAs that were encoded on the L-stand. Overall, H. suweonensis mitogenome comprised 29.2% A, 27.4% C, 14.9% G, and 28.5% T. The putative D-loop region had two tandem repeat areas, which were 160 and 121 bp in length.
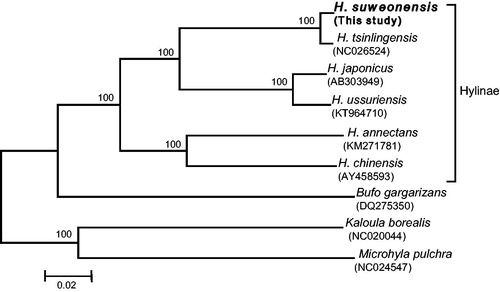
The phylogenetic relationships of the six available Hyla spp. mitogenomes were analyzed using the neighbour-joining (NJ) method in MEGA 6 (Tamura et al. Citation2013), based on 13 concatenated PCGs. This phylogenetic reconstruction showed that H. suweonensis is closer to H. tsinlingensis, contradicting previous studies (Hua et al. Citation2009; Duellman et al. Citation2016) (). The results provided here are fundamental resources for resolving Suweon tree frogs’ phylogenetic issues and for population genetic studies aiming their conservation.
Figure 1. Neighbour-joining (NJ) phylogenetic tree of six Hyla spp. frogs based on the concatenated nucleotide sequences of 13 mitochondrial protein-coding genes. Numbers at each node represent the bootstrap support value of the NJ analysis, based on 1000 replicates, and numbers below species names indicate their GenBank accession code.
Acknowledgements
We sincerely thank the one providing the specimen to the NIBR and Ju Min Jun and Sang-Hwa Lee for comments on data analyses.
Disclosure statement
The authors report no conflicts of interest and are the sole responsible for the contents and the writing of this paper.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Borzée A, Jang Y. 2015. Description of a seminatural habitat of the endangered Suweon treefrog Hyla suweonensis. Anim Cells Syst. 19:216–220.
- Chun SW, Chung EG, Voloshina I, Chong JR, Lee H, Min MS. 2012. Genetic diversity of Korean tree frogs (Hyla suweonensis and Hyla japonica): assessed by mitochondrial Cytochrome b gene and Cytochrome Oxidase Subunit I gene. Korean J Herpetol. 4:31–41.
- Duellman WE, Marion AB, Hedges SB. 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa. 19:1–109.
- Hua X, Fu C, Li J, Nieto Montes de Oca A, Wiens JJ. 2009. A Revised phylogeny of Holarctic treefrogs (genus Hyla) based on nuclear and mitochondrial DNA sequences. Herpetologica. 65:246–259.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kuramoto M. 1980. Mating calls of treefrogs (genus Hyla) in the Far East, with description of a new species from Korea. Copeia. 1:100–108.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lee JH, Park DS. 2016. Check list Organisms in Korea 17: The Encyclopedia of Korean Amphibians. p. 126.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yang SY, Min MS, Kim JB, Suh JH. 1997. Intra and inter specific diversity and speciation of two tree frogs in the genus Hyla. Gene Genom. 19:71–87.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
