Abstract
In this study, we sequenced and determined the complete mitochondrial genome of Oncorhynchus keta and Oncorhynchus masou masou, which are the economically important fish species of China. They are circular molecule of 16,656 and 16,704 bp in size, containing 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs and a displacement loop region (D-loop). Oncorhynchus keta mitochondrial genome consists of A: 27.80%, T: 26.21%, G: 16.98%, C: 29.01%. Oncorhynchus masou masou mitochondrial genome consists of A: 28.20%, T: 26.82%, G: 16.52%, C: 28.46%. The total length of the 13 protein-coding genes was 11,420 and 11,415 bp.
Oncorhynchus keta and Oncorhynchus masou masou are belong to Salmonoidea family, which are the economically important fish species in China. Oncorhynchus keta is widely distributed in North pacific and its coastal tributary, which is the typical anadromous fish (Liu et al. Citation2013). Compared to O. keta, the distritution range of O. masou masou is relatively centralized in Northwest Pacific, which occurs in two life history forms: anadromous and landlocked (Xie Citation2007). In China, the O. keta is distributed in Amur, Wusuli, Suifen Current and Tumen Rivers, and the O. masou masou is distributed in Tumen River and Suifen Current. At present, there are few reports about O. keta and O. masou masou from rivers in northeast of China (Chen Citation2005; Liu et al. Citation2013; Wang et al. Citation2013, Citation2015). Mitochondrial DNA information provided the basis for the studies in population genetic differentiation. In this study, the complete sequence of the mitochondrial genome of O. keta and O. masou masou (GeneBank accession number are KY320492 and KY320493) was discovered, which will be suitable for biogeographical and population genetic analyses of the salmons.
Oncorhynchus keta was collected from Wusuli River, which is a natural boundary between China and Russia. Oncorhynchus masou masou was collected in the middle reaches of Suifen River. The total genomic DNA was extracted with the traditional phenol-chloroform method (Taggart et al. Citation1992). Twenty-eight and 35 primers were used to amplify the target PCR products as sequencing, it is designed by primers and PCR product was purified by 3.0 Analysis ContigExpess softword (SAS Inc., Cary, NC). They are circular molecule of 16,656 and 16,704 bp in size, containing 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs and a displacement loop region (D-loop). Most of the genes are encoded on the heavy strand (H-strand), except for the eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu and tRNA-Pro) and one protein-coding gene (ND6). Oncorhynchus keta mitochondrial genome consists of A: 27.80%, T: 26.21%, G: 16.98%, C: 29.01%. Oncorhynchus masou masou mitochondrial genome consists of A: 28.20%, T: 26.82%, G: 16.52%, C: 28.46%. The total length of the 13 protein-coding genes was 11,420 and 11,415 bp.
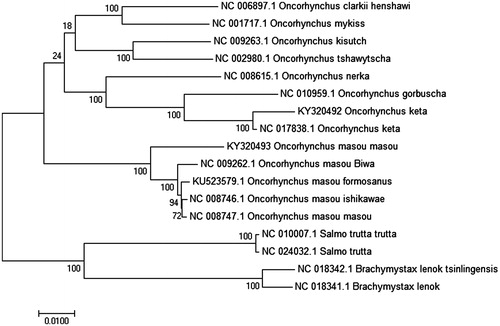
The complete mitochondrial genome sequences of 15 other closely related species are downloaded from GenBank, which were used for phylogenetic analysis (). Multiple alignments of 17 mitochondrial genomes were aligned using ClustalW. The phylogenetic tree from maximum-likelihood was reconstructed using Mega6.0 (Tamura et al. Citation2013). To obtain the confident supports, 1000 bootstrap replicates were set for the analysis. Pholygenetis analysis showed that O. masou masou shared the same cluster with other O. masou subspecies and O. keta shared the same cluster with other Oncorhynchus subspecies, they are closely relative species but they may diverge differently. We expect that the present result will help to elucidate the taxonomic status of O. masou masou and O. keta, which contribute to the conservation.
Disclosure statement
The authors declare no competing interests in the preparation and execution of this manuscript. The authors are solely responsible for its content. This research was supported by the program of Ministry of agriculture species resources protection project (Grant no. 2130135: 2010–2016).
Additional information
Funding
References
- Chen JP. 2005. Genetic analysis of four wild Chum salmon Oncorhynchus keta populations in China based on microsatellite markers. Environ Biol Fish. 73:181–188.
- Liu W, Zhan PR, Wang JL, Tang FJ. 2013. Structure of otoliths dailt growth ring of embryo Chum salmon and environmental mass marking. Acta Hydrobiol Sin. 37:929–937.
- Taggart JB, Hynes RA, Prodoh PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang JL, Liu W, Tang FJ. 2013. Analysis of biological traits of Chum salmon (Oncorhynchus keta Walbaum) in the Amur River, China. J Fish Sci China. 20:93–100.
- Wang C, Liu W, Zhan PR, Wang JL, Li PL. 2015. Exogenous Sr2+ sedimentation on otolith of Chum salmon embrys. Chin J Appl Ecol. 26:3189–3194.
- Xie YH. 2007. Freshwater Fishes in Northeast Region of China. Shenyang, China: Liaoning Science and Technology Publishing House., pp. 296–300.