Abstract
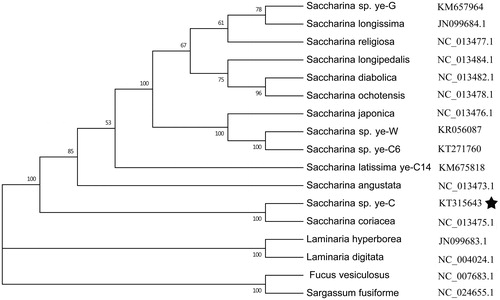
The complete sequence (37,609 bp) of the mitochondrial DNA (mtDNA) of the Saccharina sp. ye-C was determined using Illumina sequencing data. The genome contains 38 protein-coding genes (PCG), 3 ribosomal RNA (rRNA), 25 transfer RNA (tRNA) genes that are typical of Saccharina mtDNA. The phylogenetic analysis based on the mitochondrial genomes of brown algae showed that Saccharina sp. ye-C formed a robust clade with Saccharina coriacea, which strongly supports their close phylogenetic relationship.
The kelp Saccharina are large seaweeds with two distinctive generations of microscopic gametophytes and large elaborate sporophytes (Kawai et al. Citation2016). They belong to the brown algae (Phaeophyceae) in the order Laminariales. Saccharina japonica is an important economic alga which has been widely cultivated in China, Japan and Korea (Wang et al. Citation2013). However, in China S. japonica suffered close breeding for generations which have caused significant issues such as loss of genetic varieties, decreasing growth rate and massive mortality (Guan et al. Citation2016). Recently, the genome of S. japonica has been sequenced (Ye et al. Citation2015), which could strongly promote the genetic improvement of the species. In the study, genomes of several strains of Saccharina collected from different locations all over the world were re-sequenced. Under this background, we have enough Illumina sequencing data to recover some mitochondrial genomes of the re-sequenced strains to develop more reliable genetic tools. In this study, the complete mitochondrial genome of a wild strain (NO. KT315643), sampled in Sakhalin in Russia is recovered and named Saccharina sp. ye-C based on the phylogenetic analysis with 16 complete brown algae mitochondrial genomes.
Modified phenol-chloroform procedure was used to extract the total genomic DNA (Greco et al. Citation2014). 4G PE100 reads in total was produced by the second-generation Illumina sequencing.
42,452 reads were mapped to the S. japonica mitochondrial genome (37,609 in length) using Genious (http://www.genious.com) and no gap region was found in the mapping result. BLAST (Altschul et al. Citation1997), DNAstar (Burland Citation2000), and DOGMA (http://dogma.ccbb.utexas.edu) were employed to annotate the mitochondrial genome and the annotated file was submitted to NCBI using Sequin (http://www.ncbi.nlm.nih.gov/projects/Sequin/).
The length of complete Saccharina sp. ye-C is 37,609 bp and the genome contains 38 protein-coding genes (rps2-4, rps7-8, rps10-14, atp6, atp8, atp9, cox1-3, nad1-7, nad9, nad11, nad4L, rpl2, rpl5, rpl6, rpl14, rpl16, rpl19, rpl31, ORF41, ORF130, ORF377, tatC and cob), 25 transfer RNA (tRNA) genes, 3 ribosomal RNA (rRNA) genes (5S rRNA, 16S rRNA and 23S rRNA). All 38 protein-coding genes (PCGs) have typical initiation codons (ATG). The numbers of PCGs that have complete termination codons TAA, TAG, TGA are 27, 7 and 4, respectively. No incomplete stop codons were found. The overall GC content is 35.35%, which is well within the normal range of brown mitochondrial DNAs. Nucleotide frequency of the H-strand is as follows: T, 36.29%; A, 28.36%; C, 14.74%; and G, 20.61%. The mitogenome of Saccharina sp. ye-C encodes 9644 amino acids, excluding the stop codons. All the 25 typical tRNAs, ranging from 71 to 88, possess a complete clover leaf secondary structure. The rRNAs of the 5S rRNA, 16S rRNA and 23S rRNA genes are 133 bp, 1535 bp and 2736 bp in length, respectively.
Phylogenetic analysis based on other 16 brown algae complete mitochondrial sequence data show that Saccharina sp. ye-C belongs to a Saccharina clade and is closely related to S. coriacea. The result was consistent with recent phylogenetic analyses and certain morphological characters (Zhang et al. Citation2013; Xu et al. Citation2016). Complete mitochondrial genomes have enhanced the resolution and statistical confidence of inferred phylogenetic trees ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Greco M, Sáez CA, Brown MT, Bitonti MB. 2014. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the Model Brown Alga. PLoS One. 9:e96470.
- Guan Z, Fan X, Wang S, Xu D, Zhang X, Wang D, Miao Y, Ye N. 2016. Sequencing of complete mitochondrial genome of brown algal Saccharina sp. Ye-G. Mitochondrial DNA Part A. 27:2125–2126.
- Kawai H, Hanyuda T, Uwai S. 2016. Evolution and Biogeography of Laminarialean Kelps: Netherlands: Springer.
- Xu L, Wang S, Fan X, Xu D, Zhang X, Miao Y, Ye N. 2016. Sequencing of complete mitochondrial genome of brown algal Saccharina sp. ye-C6. Mitochondrial DNA Part A. 27:3733–3734
- Ye N, Zhang X, Miao M, et al. 2015. Saccharina genomes provide novel insight into kelp biology. Nat Commun. 6:6986.
- Wang WJ, Wang FJ, Sun XT, Liu FL, Liang ZR. 2013. Comparison of transcriptome under red and blue light culture of Saccharina japonica (Phaeophyceae). Planta. 237:1123–1133.
- Zhang J, Wang X, Liu C, Jin Y, Liu T. 2013. The complete mitochondrial genomes of two brown algae (Laminariales, Phaeophyceae) and phylogenetic analysis within Laminaria. J Appl Phycol. 25:1247–1253.