Abstract
Bats represent around one-fourth of the world’s mammals and their taxonomy is still controversial. Molossids are one of the most diverse bat families with a wide knowledge gap. In this study, we report the first complete mitochondrial genomes of three molossid bats: the European free-tailed bat Tadarida teniotis, the La Touche’s free-tailed bat Tadarida latouchei, and the Wrinkle-lipped free-tailed bat Chaerephon plicatus. The mitogenomes are 16,869 and 16,784 bp long for T. teniotis and T. latouchei, respectively, while in C. plicatus it is at least 16,216 bp although the control region was not fully recovered due to its higher divergence from T. teniotis. The genomes show conserved synteny with other mammalian mitogenomes, containing 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 control region (d-loop). All protein-coding genes start with the ATG start codon, except for ND2, ND3, and ND5 which begin with ATA or ATT. Eleven protein-coding genes terminated in a canonical stop codon, TAA or TAG, two contain incomplete stop codons, T or TA. Cytochrome b terminates in the mitochondria-specific stop codon AGA. These mitogenomes provide a valuable resource for future studies of Molossidae and other bat and mammal species.
Molossidae is the fourth largest bat family (Mammalia, Chiroptera), with ∼100 species divided into 17 genera (IUCN Citation2017). Although molossids are distributed throughout tropical and subtropical regions of the world, the range of most species is poorly known. Many species are similar in appearance and difficult to capture due to their fast and high-flying behaviours (Vaughan Citation1966). This has led to an under-representation of this family in museum collections and several taxonomic inconsistencies (Ammerman et al. Citation2012). In the Eurasian region there are at least three recognized species of free-tailed bats: Tadarida teniotis complex (composed of Tadarida teniotis, Tadarida insignis, Tadarida latouchei, and Tadarida coecata), Tadarida aegyptiaca and Chaerephon plicatus (Hutson et al. Citation2001). The teniotis species group has been subject to great debate, with some authors considering insignis, latouchei and coecata as subspecies of teniotis, while more recently others consider insignis and latouchei as full species, and coecata as a subspecies of insignis (Simmons Citation2005). Morphological analysis of teniotis, insignis, and latouchei, have found consistent differences, suggesting the full species status for each group, and therefore the restriction of teniotis to the west of India, and insignis to further east (Funakoshi & Kunisaki Citation2000). However, to date this classification has lacked any molecular support. In fact, there are no available mitogenomes for any species of the family Molossidae. Our results provide useful reference for future phylogenetic and phylogeographic studies.
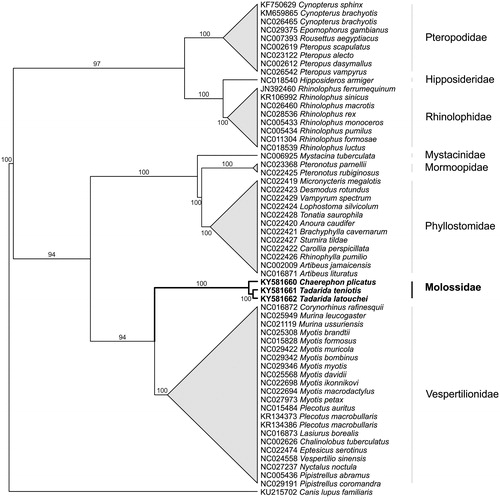
Genomic DNA was extracted from T. teniotis (Portugal, 41.287 N 6.873 W), T. latouchei (Laos, 20.153 N 103.407 E, voucher ROM MAM 118321), and C. plicatus (Laos, 20.723 N 101.138 E, voucher ROM MAM 118373) tissue samples using QIAamp DNA Micro Kit (QIAGEN). Due to the degraded state of T. latouchei and C. plicatus samples, a mitochondrial capture-based protocol was used. Specific primers for T. teniotis were designed in order to amplify 2 overlapping fragments of ∼10kb (MtF13 (5′-TGCATTACACATCCGACACA-3′) with MtR12 (5′-GGCTTTGAAGGTCCTTGGTC-3′), and MtF12 (5′-CGGCTAACATACGCTACATCC-3′) with MtR13 (5′-GCCTATGAAGGCAGTGGCTA-3′) using Takara LA Taq polymerase. Custom primers had to be designed due to several problems in the amplification process related to nuclear copies of mitochondrial genes. Complete mitochondrial mitogenomes of each sample were then captured following the protocol by Maricic et al. (Citation2010) and sequenced with 250 bp paired end reads in Illumina MiSeq (Vairão, Portugal). Due to the high divergence between T. teniotis and C. plicatus, the control region d-loop of C. plicatus mitochondria was not fully recovered. The mitogenomes (Genbank accession no. KY581660/61/62) were assembled de novo and annotated using Geneious 9.1.5 (Kearse et al. Citation2012). A neighbour-joining tree was built with the Tamura–Nei distance to reconstruct the phylogeny of the molossidae family using Geneious based on an alignment of 55 bat species mitogenomes (without d-loop) plus the ones from this study (Saitou & Nei Citation1987).
Molossidae were more closely related to Vespertilionidae (), with Tadarida species forming a separate clade from Chaerephon. Average coding gene distance between T. teniotis and T. latouchei and C. plicatus was 10% and 15%, respectively. These results suggest that T. latouchei is indeed a different species from T. teniotis, although nuclear markers would be needed to confirm this. Molossidae are one of the most diverse family of bats, however these are the first mitogenomes available. Future studies with more taxa and molecular markers are needed to better understand the phylogeny and the taxonomic status of this highly speciose family of mammals.
Acknowledgements
The study was funded by Fundacão para Ciência e Tecnologia (FCT) (Project LTER/BIA-BEC/0004/2009) and EDP Biodiversity Chair. FCT funded Vanessa Mata (PD/BD/113462/2015), Francisco Amorim (PD/BD/52606/2014) and Hugo Rebelo (IF/00497/2013). Support has also been received from the EU FP7 for research, technology development and demonstration (Grant agreement No 286431) and the EU Horizon 2020 research and innovation programme (Grant agreement No 668981). The authors would like to thank Joana Rocha for all the support in the lab.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Ammerman LK, Lee DN, Tipps TM. 2012. First molecular phylogenetic insights into the evolution of free-tailed bats in the subfamily Molossinae (Molossidae, Chiroptera). J Mammal. 93:12–28.
- Funakoshi K, Kunisaki T. 2000. On the validity of Tadarida latouchei, with reference to morphological divergence among T. latouchei, T. insignis and T. teniotis (Chiroptera, Molossidae). Mammal Study. 25:115–123.
- Hutson AM, Mickleburgh SP, Racey PA. (comp.). (2001) Microchiropteran bats: global status survey and conservation action plan. IUCN/SSC Chiroptera Specialist Group. Gland, Switzerland and Cambridge (UK): IUCN.
- IUCN. 2017. The IUCN red list of threatened species. Version 2017-2 [Internet]. [cited 2017 Feb 01]. Available from: http://www.iucnredlist.org/
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Maricic T, Whitten M, Pääbo S. 2010. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 5:9–13.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Simmons NB. 2005. Order Chiroptera. In: Wilson DE and Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd edition. Baltimore: The Johns Hopkins University Press; p. 312–529.
- Vaughan TA. 1966. Morphology and flight characteristics of molossid bats. J Mammol. 47:249–260.