Abstract
In this study, combining the liver transcripts from both sexes by RNA-Seq with DNA sequences by conventional PCRs, we have determined the complete mitogenome of Hainan medaka Oryzias curvinotus collected from the mangrove seawater of the Leizhou Peninsula in tropical South China (Accession no.: KY364884). The mitochondrial genome is 16,676 bp, and its content and structure are highly homologous to those of other teleostean fishes, including 13 protein-coding genes (PCGs), 2 rRNAs, 22 tRNA and 1 control region. Among the PCGs, ATG is used as the initiation codon, except for GTG in COI gene. There are 7 overlapping genes with overlap lengths ranging from 1 to 10 nucleotides (nt), while ten intergenic regions with a total of 66 nt and a maximum interval of 37 nt between tRNAAsn and tRNACys. Moreover, the data from RNA-Seq shows that the significant differences exist in the expression patterns of mitogenomes between male and female.
Hainan medaka Oryzias curvinotus (Teleostei: Beloniformes) is a potential model fish akin to the freshwater Japanese medaka (Oryzias latipes), mainly distributed along the northwest coast of the South China Sea (Parenti Citation2012; Ta & Tran Citation2016). However, the mitochondrial genome of O. curvinotus was still not determined.
Here, the mature fishes were collected from National Mangrove Nature Reserve in Leizhou Peninsula, Guangdong Province, China. The muscle was used to extract DNA (Guo et al. Citation2016), while the livers of females and males were used to purify total RNA for transcriptomic sequencing (RNA-Seq) (Wang et al. Citation2009). The typical specimen and DNA were deposited in the Guangdong Ocean University. According to the sequences of RNA-Seq, three pairs of primers for conventional PCR were designed. The amplified DNA products were sequenced to fill the gaps and substitute the 3′ transcript ends between the Cytb gene and the control region, the control region and the 12s rRNA gene, the 12S rRNA gene and the 16S rRNA gene.
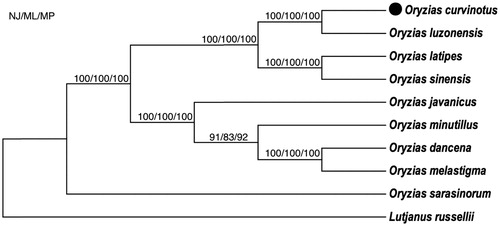
As a result, we have determined the mitogenome of Oryzias curvinotus. Based on the complete mtDNA, as shown in , the phylogenetic trees supported the genetic relationship between the O. curvinotus and the O. luzonensis is the closest among the 7 species of genus Oryzias: O. curvinotus, O. luzonensis, O. latipes, O. minutillus, O. javanicus, O. sinensis and O. melastigma (also O. dancena), which is consistent with the previous reports using the nuclear tyrosinase and mitochondrial 12S and 16S rRNA genes (Naruse Citation1996; Takehana et al. Citation2005).
Figure 1. Neighbor-joining (NJ), maximum-likelihood (ML) and maximum-parsimony (MP) consensus tree based on mitogenomic DNA sequences. All the bootstrap values are indicated at the nodes.
Furthermore, through mapping the clustered transcripts to the annotated mitogenome of O. curvinotus, the obvious differences of the transcript units and their expression levels were observed between both sexes. Within a sex, the expression levels were also various among the transcripts. Torres et al. (Citation2009) reported a similar result found in drosophila. Recently, more researches indicated that mitogenomic transcripts should be more complicated processing than previously assumed (Wang et al. Citation2013; Perera et al. Citation2016).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. This project was funded by the grants from the National Natural Science Foundation of China (NSFC) (No: 31201996), and Excellent Young Teachers Program of Guangdong Province (Yq2013091 and Yq2013089).
Additional information
Funding
References
- Guo Y, Bai Q, Yan T, Wang Z, Liu C. 2016. Mitogenomes of genus Pristipomoides, Lutjanus and Pterocaesio confirm Caesionidae nests in Lutjanidae. Mitochondrial DNA A DNA Mapp Seq Anal. 27:2198–2199.
- Naruse K. 1996. Classification and phylogeny of fishes of the genus Oryzias and its relatives. Fish Biol J Medaka. 8:1–9.
- Parenti LR. 2012. Oryzias curvinotus. in The IUCN Red List of Threatened Species.
- Perera OP, Walsh TK, Luttrell RG. 2016. Complete mitochondrial genome of Helicoverpa zea (Lepidoptera: Noctuidae) and expression profiles of mitochondrial-encoded genes in early and late embryos. J Insect Sci. 16:1–10.
- Ta TT, Tran HD. 2016. Dependence of Hainan medaka, Oryzias curvinotus (Nichols & Pope, 1927), on salinity in the Tien Yen estuary of Northern Vietnam. Anim Biol. 66:49–64.
- Takehana Y, Naruse K, Sakaizumi M. 2005. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol Phylogenet Evol. 36:417–428.
- Torres TT, Dolezal M, Schlotterer C, Ottenwalder B. 2009. Expression profiling of Drosophila mitochondrial genes via deep mRNA sequencing. Nucleic Acids Res. 37:7509–7518.
- Wang HL, Yang J, Boykin LM, Zhao QY, Li Q, Wang XW, Liu SS. 2013. The characteristics and expression profiles of the mitochondrial genome for the Mediterranean species of the Bemisia tabaci complex. BMC Genomics. 14:401.
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
