Abstract
The complete chloroplast genome of the microalgae Dunaliella salina strain SQ was determined in this study. The total length of the chloroplast genome is 243,635 bp with 29.73% GC content. The genome is composed by a small single copy (SSC) region of 101,527 bp and a large single-copy region of 107,815 bp separated by two inverted repeats (IR) regions of 17,145 bp. A total of 98 genes were annotated, including 66 coding genes, 3 rRNAs, and 29 tRNAs. This complete plastid genome can be used to elucidate genetic variations in organellar genomes between D. salina strains.
In the past decade, green microalgae have gained interest as a source of different biocompounds such as carotenoids and other molecules of biotechnological interest. Dunaliella salina is a unicellular green microalgae known for been halotolerant and one of the richest sources of natural β-carotene (Lamers et al. Citation2008). There are well-known beneficial properties of carotenes in human health, extracts made from D. salina have shown anti-proliferative, anti-inflammatory, and cytotoxic activity against certain types of cancer, these properties may be due to the high content of carotenes (Chiu et al. Citation2016; Singh et al. Citation2016). Previously, the chloroplast and mitochondrial genomes of D. salina CCAP 19/18 has been sequenced by Smith et al. (Citation2010) and the mitochondrial genome of D. salina CONC-001 has also been described (Del Vasto et al. Citation2015); the mitochondrial genomes present several differences between strains. Here, we report the complete chloroplast genome of D. salina SQ to provide information about the genetic variation between organellar genomes in D. salina strains.
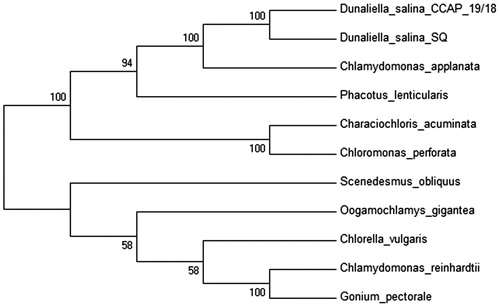
Dunaliella salina was collected from San Quintin in Baja California, México (30° 32′ 13.91″ N, 116° 2′ 1.22″ W) from a hypersaline lagoon. A single cell was isolated and cultured in modified liquid medium (Feng et al. Citation2009) with 250 mM NaCl. Total chloroplast DNA was extracted with AxyPrep Multisource Genomic DNA Miniprep Kit (Axygen Biosciences, Union City, CA) and pure cpDNA was sequenced using Illumina MiSeq (UC, San Diego, CA). The genome was de novo assembled using A5 miseq pipeline (Coil et al. Citation2014) and Ray v2.3.2 (Boisvert et al. Citation2010). Annotation of the cpDNA was performed with RNAweseal (Lang et al. Citation2007), MFannot (Beck & Lang Citation2010) and tRNAscan-SE 1.21 (Schattner et al. Citation2005) and the results were validated with BLAST searches. The complete annotated chloroplast genome was submitted to GenBank to obtain the accession number KX530454. The complete chloroplast genome has a total length 243,635 bp with 29.73% of GC content and included two inverted repeats (IR’s) of 17,145 bp each separated by a large single-copy (LSC) region of 107,815 bp and a small single-copy (SSC) region of 101,527 bp. The genome contains 98 genes, including 66 protein-encoded genes, 29 tRNA genes and 3 rRNA genes. Five genes are situated in the IR region (rrns, rrnl, rrn5, trnA, and trnI). Eight genes contain introns, one with five introns (psbA), one with four introns (rrnL), two with two introns (psbC and rrns), and three contain a single intron (atpA, psaB, and psbC). The psaA gene is trans-spliced, the first two exons are situated in the LSC region while the remaining three exons are located in the SSC region and two introns were detected between them. For the phylogenetic analysis, 10 chloroplast genomes sequences from others microalgae where selected and downloaded from NCBI database and a neighbour-joining tree with 500 bootstraps was constructed using MEGA7 (Kumar et al. Citation2016) ().
Figure 1. Phylogenetic tree of D. salina SQ and 11 microalgae chloroplast genomes including another strain of D. salina. The GeneBank accession numbers are listed as follow: Characiochloris acuminata (KT625418), Chlamydomonas reinhardtii (NC_005353.1), C. applanata (KT625417), Chlorella vulgaris (NC_001865.1), Chloromonas perforata (KT625416), D. salina (NC_016732), D. salina strain SQ (KX530454) (this study), Gonium pectoral (AP012494), Oogamochlamys gigantean (KT625412), Phacotus lenticularis (KT625422) and Scenedesmus obliquus (DQ396875).
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Beck N, Lang B. 2010. MFannot, organelle genome annotation webserver [Internet]. Available from: http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17:1519–1533.
- Chiu HF, Liao JY, Lu YY, Han YC, Shen YC, Venkatakrishnan K, Golovinskaia O, Wang CK. 2016. Anti‐proliferative, anti‐inflammatory and pro‐apoptotic effects of Dunaliella salina on human KB oral carcinoma cells. J Food Biochem e12349.
- Coil D, Jospin G, Darling AE. 2014. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587--589.
- Del Vasto M, Figueroa-Martinez F, Featherston J, Gonzalez MA, Reyes-Prieto A, Durand PM, Smith DR. 2015. Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biol Evol. 7:656–663.
- Feng S, Xue L, Liu H, Lu P. 2009. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol Biol Rep. 36:1433–1439.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870--1874.
- Lamers PP, Janssen M, De Vos RC, Bino RJ, Wijffels RH. 2008. Exploring and exploiting carotenoid accumulation in Dunaliella salina for cell-factory applications. Trends Biotechnol. 26:631–638.
- Lang BF, Laforest MJ, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet. 23:119–125.
- Singh P, Baranwal M, Reddy SM. 2016. Antioxidant and cytotoxic activity of carotenes produced by Dunaliella salina under stress. Pharm Biol. 54:2269–2275.
- Smith DR, Lee RW, Cushman JC, Magnuson JK, Tran D, Polle JE. 2010. The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 10:83.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
