Abstract
Phylogenetic analyses of nuclear and mitochondrial genomes indicate that polar bears captured the brown bear mitochondrial genome 160,000 years ago, leading to an extinction of the original polar bear mitochondrial genome. However, mitochondrial DNA occasionally integrates into the nuclear genome, forming pseudogenes called numts (nuclear mitochondrial integrations). Screening the polar bear genome identified only 13 numts. Genomic analyses of two additional ursine bears and giant panda indicate that all except one of the discovered numts entered the bear lineage at least 14 million years ago. However, short read genome assemblies might lead to an under-representation of numts or other repetitive sequences. Our findings suggest low integration rates of numts in bears and a loss of the original polar bear mitochondrial genome.
Polar and brown bears are two well-recognized species, which differ in their morphology and ecology (Nowak Citation1999). Recent research has shown that polar bears diverged from brown bears in the mid Pleistocene (Hailer et al. Citation2012; Liu et al. Citation2014), which is supported by the fossil record (Kurtén & Anderson Citation1980) (but see Miller et al. (Citation2012) for earlier divergence estimates). Phylogenetic analyses of mitochondrial DNA (mtDNA) showed that polar bears appear to be nested inside the brown bear radiation and dated the emergence of polar bears around 160 thousand years ago (kya) (Lindqvist et al. Citation2010; Edwards et al. Citation2011; Hailer et al. Citation2012). The deviating mtDNA phylogeny can be explained by recurrent introgressive hybridization between female brown bears and male polar bears 160 kya, resulting in a mitochondrial capture event that replaced the original polar bear mitochondrial (mt) genome (Miller et al. Citation2012). As evident from the low genetic diversity among polar bears, population bottlenecks during interglacials likely led to a fixation of the introgressed brown bear mtDNA in the polar bear lineage and a loss of the original polar bear mtDNA.
Occasionally mt genome sequences are transferred to the nucleus, and are incorporated as pseudogenes called numts (nuclear sequence of mitochondrial origin, pronounced ‘new-mite’) (Lopez et al. Citation1994, Rogers & Griffith-Jones Citation2012). Numt insertions occur via non-homologous end joining at double-strand breaks in the nuclear genome. In general, the genomic fraction of numts is less than 0.1% with the highest proportion found in plants and yeast (0.28%) (Hazkani-Covo et al. Citation2010). Several genome scale studies have discovered that the total copy number and sequence length of numts varies widely between mammalian species (Hazkani-Covo et al. Citation2010). For instance, only 49 copies, totalling 6 kilo base pairs (kb), are found in the rat (Rattus norvegicus) genome while 1859 copies (2093 kb) are found in opossum (Monodelphis domestica) (Hazkani-Covo et al. Citation2010). Three different processes can contribute to the differences in the number of numts between species: the frequency of mitochondrial transfer, the amount of integrations as well as the dynamics of insertion processes.
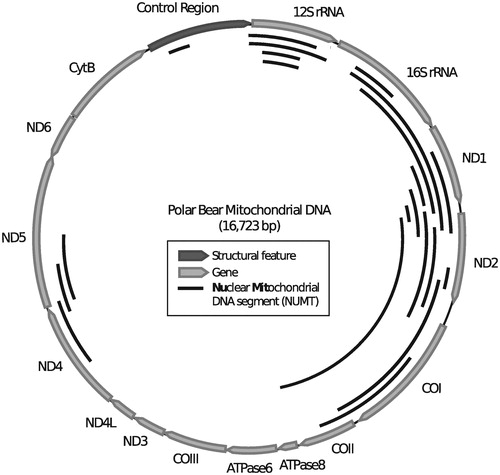
To date numt insertions have not been studied in representatives of the bear family (Ursidae). If indeed, at 160 kya, the polar bear mt genome was replaced by the brown bear mt genome, all polar bear numts that entered the nuclear genome between the divergence of both species (∼600 kya) and the mitochondrial capture event (∼160 kya) are genomic ‘fossils’ of the original polar bear mtDNA (). We screened the polar bear genome for numts to reconstruct the ancient polar bear mt genome sequence. Although the rate of numt integration in the genome is generally low, polymorphic numt copies are known from the human population (Dayama et al. Citation2014), indicating that these polymorphisms have spread within a few hundred generations. Screening the polar bear genome sequence, using the mt genome found in extant polar bears, identified 64 putative numts totalling about 29 kb sequence. The identified numt sequences cover 62% of a bear mt genome, i.e. 38% of the mt genome did not contribute to the polar bear numt landscape. We focused our analyses on 22 numts that were longer than 200 base pairs (bp), which represent 57% of the mt genome and represent mainly the NADH1 to COII region. Both rRNA genes as well as NADH4 and NADH5 are only partially covered (). Phylogenetic maximum likelihood analysis of the numt sequences, including homologous mitochondrial sequences of all living bear species and other carnivores, indicated that the numts are older than 14 million years (My) (Supplementary Data 1). Fourteen of the 22 identified numts form clusters consisting of two or three numt fragments localized on the same scaffold (Supplementary Table 2). For each cluster, the scaffold localization, the gene order and orientation suggest that these are eroded fragments from single longer ancient integrations. Merging the consecutive fragments in a cluster reduced the total number to 13 numts (Supplementary Table 2). Aligning the genomic polar bear numt loci to orthologous sequences from the giant panda (Ailuropoda melanoleuca) (Li et al. Citation2010a) revealed that 11 of the 13 numts are present in full length in the giant panda genome and thus integrated at least 14 My ago, i. e. before the giant panda lineage diverged, (Kumar et al. Citation2017). Locus 7 was only partially identified and locus 8 was not identified in the giant panda genome. The genomic distance between numt fragments in the polar bear genome matched approximately the distance of the same fragments when mapped to the mt genome (Supplementary Table 3). Numts were fragmented by interspersed transposable elements (loci 7, 9 and 10) or expanding short tandem repeats (loci 6 and 11). Some of these interspersed sequences were only found in polar bear or giant panda, causing different inter-fragment distances (Supplementary Table 3). A full LINE1-1_Ame transposable element was inserted in locus 10 in the giant panda genome. Nearly all identified polar bear numt sequences seem to have been inserted prior to the evolution of Ursidae and no numts representing ancestral or recent polar bear mtDNA have entered the nuclear genome (). As an additional line of evidence for decreased numt integration in Ursidae, we analyzed whole-genome sequencing data of two additional bear species for polymorphic numt insertions using the structural variation (SV) caller Lumpy (Layer et al. Citation2014). Our SV analyses date the previously identified numt insertions to at least before the diversification of American black (Ursus americanus), brown and polar bear, which gives a minimum age of 3 My (Kumar et al. Citation2017). However, the comparative screening of the giant panda genome indicate a much older numt insertion for most of the loci.
Figure 1. Phylogeny of bears reconstructed by nuclear DNA (nuDNA, left side) and mtDNA (right side). The left phylogeny reflects the speciation history of bears. About 160 kya, the original polar bear mtDNA lineage was replaced by brown bears (dashed arrow) causing the observed paraphyly of brown bears in the mtDNA phylogeny (right side). Dashed lines above the nuDNA phylogeny indicate the timeframe for potential integration of numts that represent the original polar bear mtDNA (PB-numts) and the observed reduction of numt integration in Ursidae.
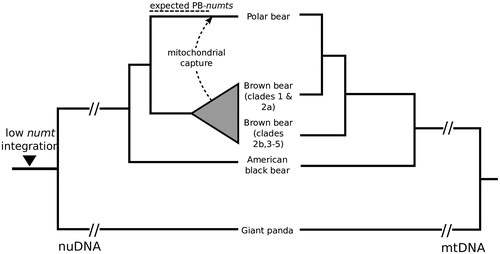
Figure 2. Genetic map of the polar bear mitochondrial genome with annotated genes. The identified numts longer than 200 bp are shown as solid black lines on the inside of the mt genome.
The number of numts and the genomic fraction derived from numts are not unreasonably low in the polar bear genome when compared to other mammals, that show similar low numt fractions. For example, the two murid rodents, mouse (Mus musculus) and rat only have between 6-39 kb numts in their respective genomes, which is among the lowest numbers of numts in mammals (Hazkani-Covo et al. Citation2010). The low incidence of numts in the closely related mouse and rat, as well as in the bear family suggest that these groups may have evolved mechanisms to suppress the integration of numts or that the rate of deletion is higher than in other groups (Hazkani-Covo et al. Citation2010).
However, another important reason for the lack of recent numts in the polar bear genome sequence may lie in the genome assembly processes. Recent numts are genetically similar to the mitochondrial genome, and de Bruijn graph based assembling algorithms might not be able to distinguish between short reads originating from numts or mtDNA (Li et al. Citation2010b; Hahn et al. Citation2013). This would cause a severe under-representation of recently integrated numts in short-read-based genome assemblies, and may in addition have introduced a bias in numerous published genome assemblies. So-called, third generation sequencing technologies like PacBio or Nanopore produce long reads, that are more likely to span complete numt insertion(s) and thus facilitate their incorporation into genome assemblies (Sohn & Nam Citation2016). To date, the gorilla genome is the only non-hominid mammalian genome generated by extensive usage of PacBio sequences (Gordon et al. Citation2016), however several additional long-read-based genome assemblies will likely become available in the next years. Thus, long-read-based sequencing of a bear genome can give further insights into the evolution of numts in Ursidae.
Recently inserted numts create problems for population and phylogenetic analyses of mt genes, if they are PCR amplified instead of the mt genes (Bensasson et al. Citation2001), but appear absent from the bear family. Thus, previous mitochondrial studies of bear phylogeny and population structures (Edwards et al. Citation2011) would be free from artefacts in the form of mitochondrial pseudogenes.
Conclusions
The current polar bear genome assembly lacks recently inserted numts. The majority of the identified numt insertions have a minimum age of 14 My. A low numt insertion frequency has been reported for rodents and might be common in other mammalian groups. Thus, a low number of numts in bear genomes is not unreasonable, but an artefact from whole genome assembly algorithms cannot be excluded until further de novo assembled bear genomes become available. Utilizing long-read high-throughput technologies like PacBio or Nanopore might yield further insight into the fate of numts in Ursidae or other taxonomic groups with suspected numt depletion.
TMDN_A_1318673_Supplementary_I_nformation.pdf
Download ()Acknowledgments
The authors thank S. Gallus for creating the mitochondrial genome annotation figure. The publication of this article was funded by the Open Access Fund of the Leibniz Association.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bensasson D, Zhang D-X, Hartl DL, Hewitt GM. 2001. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 16:314–321.
- Dayama G, Emery SB, Kidd JM, Mills RE. 2014. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 42:12640–12649.
- Edwards CJ, Suchard MA, Lemey P, Welch JJ, Barnes I, Fulton TL, Barnett R, O’Connell TC, Coxon P, Monaghan N, et al. 2011. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol. 21:1251–1258.
- Gordon D, Huddleston J, Chaisson MJP, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Dunn C, et al. 2016. Long-read sequence assembly of the gorilla genome. Science (NY). 352:aae0344.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hailer F, Kutschera VE, Hallström BM, Klassert D, Fain SR, Leonard JA, Arnason U, Janke A. 2012. Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science. 336:344–347.
- Hazkani-Covo E, Zeller RM, Martin W. 2010. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genetics. 6:e1000834.
- Kolokotronis S-O, Macphee RDE, Greenwood AD. 2007. Detection of mitochondrial insertions in the nucleus (numts) of pleistocene and modern muskoxen. BMC Evol Biol. 7:67.
- Kumar V, Lammers F, Bidon T, Pfenniger M, Kolter L, Nilsson MA, Janke A. 2017. The evolutionary history of bears is characterized by gene flow across species. Scientific Reports. 7:46487.
- Kurtén B, Anderson A. 1980. Pleistocene mammals of North America. J Mammal. 62:653–654.
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 15:R84.
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. 2010a. The sequence and de novo assembly of the giant panda genome. Nature. 463:311–317.
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. 2010b. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20:265–272.
- Lindqvist C, Schuster SC, Sun Y, Talbot SL, Qi J, Ratan A, Tomsho LP, Kasson L, Zeyl E, Aars J, et al. 2010. Complete mitochondrial genome of a pleistocene jawbone unveils the origin of polar bear. Proc Nat Acad Sci USA. 107:5053–5057.
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell. 157:785–794.
- Lopez JV, Yuhki N, Masuda R, Modi W, Brien SJO. 1994. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 39:174–190.
- Miller W, Schuster SC, Welch AJ, Ratan A, Bedoya-Reina OC, Zhao F, Kim HL, Burhans RC, Drautz DI, Wittekindt NE, et al. 2012. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc Natl Acad Sci. 109:E2382–E2390.
- Nowak RM. 1999. Walker’s mammals of the world. 6th ed. Baltimore: Johns Hopkins University Press.
- Rogers HH, Griffith-Jones S. 2012. Mitochondrial pseudogenes in the nuclear genomes of drosophila. PLoS One. 7:e32593.
- Sohn J-i, Nam J-W. 2016. The present and future of de novo whole-genome assembly. Brief Bioinformatics. October, bbw096.
