Abstract
We present the first mitochondrial genome sequence of the great hammerhead shark, Sphyrna mokarran. This species is of considerable conservation concern throughout its global distribution, and currently listed as Endangered on the IUCN Red List. The mitochondrial genome is 16,719 bp in length with 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a non-coding control region. The gene arrangement is congruent with other shark and most vertebrate species. This S. mokarran mitogenome provides a genomic resource for assisting with population studies and conservation efforts for this highly depleted species.
The great hammerhead shark, Sphyrna mokarran (Sphyrnidae), is an elasmobranch of considerable conservation concern (Endangered: International Union for the Conservation of Nature (IUCN) Red List) (Denham et al. Citation2007). The slow life-history traits of this shark coupled with commercial overexploitation for its high value fins has resulted in substantial population declines throughout much of its distribution. Due to the slower evolutionary rate of mitochondrial genes in elasmobranchs compared to other vertebrates, complete mitochondrial genome sequences can offer greater resolution than single gene studies when examining the population structure and phylogeny of elasmobranchs (Feutry et al. Citation2014; Martin et al. Citation1992). This higher resolution can be critical in elucidating evolutionary significant units for management and conservation purposes (Feutry et al. Citation2015). Here we present the first characterization of the whole mitochondrial genome of the great hammerhead shark to assist in future population genetic, conservation and phylogenetic assessments.
The specimen sequenced was a male shark sampled in 2010 off the Florida Keys, USA (geospatial coordinates: 24.36667, −82.41666). A fin clip sample (accession number OC-226) is stored in 100% ethanol at Nova Southeastern University, College of Natural Sciences and Oceanography. DNA was extracted from the fin clip using a QIAGEN DNeasy Blood & Tissue Extraction Kit (Qiagen Inc., Valencia, CA) and amplified in four fragments via long PCR using KAPA HiFi HotStart ReadMix PCR (Kapa Biosystems, Boston, MA). Amplicons were purified with QIAGEN QIAquick Gel Extraction Kit and Agencourt® AMPure XP beads (Beckman Coulter Life Sciences, Indianapolis, IN). Libraries were prepared for sequencing on an Illumina MiSeq system with paired-end reads (2x250 bp) using a Nextera XT DNA Library Preparation Kit. Fastq files were trimmed using the Minoche protocol (Eren et al. Citation2013). A de novo assembly was created with Velvet 1.1 (Zerbino Citation2010), and the single ambiguity seen resolved by performing a subsequent assembly with Bowtie2 (Langmead & Salzberg Citation2012) using the consensus sequence from Velvet as the reference index. This final assembly was annotated using MitoAnnotator (Iwasaki et al. Citation2013).
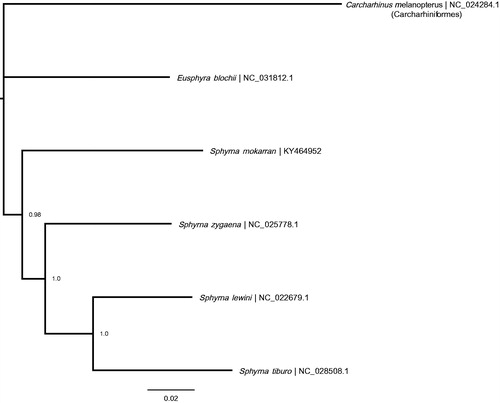
The mitochondrial genome of the great hammerhead shark (gb: KY464952) is 16,719 bp long containing 13 protein-coding, 22 tRNA, and 2 rRNA genes, and 1 non-coding control region. The nucleotide base composition is: 31.6% A, 25.6% C, 13.0% G, 29.8% T. The COI gene began with the alternative start codon GTG. The COII and ND4 genes contained incomplete stop codons. A MUSCLE alignment was performed in Geneious® 7.1.9 (http://geneious.com, Kearse et al. Citation2012) with the four other published Sphyrnidae mitochondrial genomes (Eusphyra blochii: NC_031812.1, Sphyrna lewini RefSeq: NC_022679.1, Sphyrna tiburo RefSeq: NC_028508.1, Sphyrna zygaena RefSeq: NC_025778.1), using the blacktip reef shark, Carcharhinus melanopterus, mitochondrial genome as the outgroup (RefSeq: NC_024284.1). The order of genes on the great hammerhead mitochondrial genome is congruent with the other Sphyrnidae sharks. A Bayesian tree () was created with default parameters in MrBayes 3.2 (Huelsenbeck & Ronquist Citation2001; Ronquist & Huelsenbeck Citation2003) and the best substitution model (GTR + I) determined by the Bayesian Information Criterion in JModelTest2.1.7 (Darriba et al. Citation2012).
Acknowledgements
We thank S. Gulak and the NMFS Panama City Observers Program for the shark specimen. We thank Dr. J. Lopez and J. Skutas of the Nova Southeastern University Microbiology and Genetics Lab for use of their MiSeq System.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and high-performance computing. Nat Methods. 9:772.
- Denham J, Stevens J, Simpfendorfer CA, Heupel MR, Cliff G, Morgan A, Graham R, Ducrocq M, Dulvy ND, Seisay M, et al. 2007. Sphyrna mokarran. The IUCN Red List of Threatened Species. 2007:e.T39386A10191938.
- Eren AM, Vineis JH, Morrison HG, Sogin ML. 2013. A filtering method to generate high quality short reads using Illumina paired-end technology. PLoS One. 8:e66643.
- Feutry P, Kyne PM, Pillans RD, Chen X, Marthick JR, Morgan DL, Grewe PM. 2015. Whole mitogenome sequencing refines population structure of the Critically Endangered sawfish Pristis pristis. Mar Ecol Prog Ser. 533:237–244.
- Feutry P, Kyne PM, Pillans RD, Chen X, Naylor GJ, Grewe PM. 2014. Mitogenomics of the Speartooth Shark challenges ten years of control region sequencing. BMC Evol Biol. 14:232.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Martin AP, Naylor GJP, Palumbi SR. 1992. Rates of mitochondrial DNA evolution in sharks are slow compared with mammals. Nature. 357:153–155.
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Sphyrna mokarran. [cited 2017 Apr 19]. Available from: http://dx.doi.org/10.2305/IUCN.UK.2007.RLTS.T39386A10191938.en
- Zerbino DR. 2010. Using the velvet de novo assembler for short‐read sequencing technologies. Curr Protocols Bioinform. 31:11.5.1–11.5.12.