Abstract
As an unclassified species, white-type Channa argus has existed for many years. In the present study, the relationships and genetic diversity in two colour morphs of northern snakehead were determined by the analysis of mitochondrial 12S rRNA gene, as well as other Channa species. The results of sequence analysis showed that the average genetic distance was 0.046 with the interspecies genetic distance ranging from 0.000 to 0.130; and the average interspecies genetic distance was estimated as 0.003 (range: 0.000–0.006). For the two colour morphs of Channa argus, the mean pair-wise genetic distance between them was also estimated as 0.003. Moreover, molecular phylogenetic tree was constructed using MEGA6.0, which showed that all the haploids gathered together as a branch and crossed each other. These results indicated that the white and biocolour-type Channa argus belonged to the same species, rather than subspecies at the molecular level, and the white-type Channa argus should be regard as an albino of biocolour type.
Keywords:
Introduction
The northern snakeheads C. argus (Perciformes, Channoidei, Channidae), generally dark green with large black blotches, is widely distributed in tropical and subtropical lakes, reservoirs, rivers, streams, ponds, ditches, marshes, and other fresh waters of Asia and Africa (Berra Citation2007; Bhat et al. Citation2014). The white-type C. argus is only discovered in the Jialing River in Sichuan (105.05E, 29.58N) in China, which is white without any blotches and has a higher value for development, as well a potential ornamental value. In recent years, two distinct colour morphs of northern snakehead were successively discovered in China () (Ding Citation1994; Zhou et al. Citation2015).
Figure 1. Molecular phylogenetic tree constructed by NJ method based on 12S rRNA gene using Kimura 2-parameter model (high bootstrap values (>80%) in 1,000 resamplings are shown at the corresponding nodes).
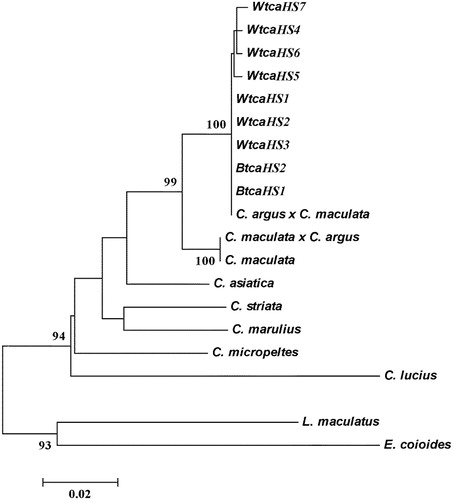
The two colour morphs have been regarded as the distinct species from C. argus by researchers (Kimura Citation1934; Su & Xiong Citation2001). However, some others have considered the two colour morphs as colour variations of one single species (Wang et al. Citation1992, Citation1993). These different classifications in two colour morphs were based solely on morphological aspects. In order to determine the relationship and genetic diversity of two colour morphs of northern snakehead, the mitochondrial 12S rRNA sequences were used as molecular markers. Our aim was to provide crucial genetic information for the appropriate classification in white-type C. argus.
Materials and methods
Sample collection and DNA extraction
All samples were identified according to morphological characters (Cheng & Zheng Citation1987; Courtenay & Williams Citation2004). Basic information of sampling sites and size (n) of Channa species are given in . Total genomic DNA was extracted from the caudal fin using a standard DNA extraction kit (DNeasy tissue kit, Baitaike Biotech Co., Ltd, Beijing, China). The amplifying primers were as follows: 12S-F:AAGGTTTGGTCCTGACTTTA CT; 12S-R ACTTACCATGTTA-CGACTTGCC. The PCR amplification was carried out in 50 μl volumes consisting of 25 μl of 2 × PCR mix buffer and 0.5 μl of 2.5U/μl Taq DNA polymerase (Dongsheng Biotech Co., Ltd, Guangzhou, China), 2 μl of 100 ng/μl DNA template, 2 μl of 10 mM each primer and 20.5μl of sterile ultrapure water. Thermal cycling condition consisted of 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, annealing at 55 °C for 50 s, and an extension temperature of 72 °C for 90 s, and then followed by a final extension of 72 °C for 10 min. The PCR product was purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Sequencing was conducted on an ABI377 automatic sequencer with both forward and reverse primers.
Table 1. Basic information of sampling sites and size (n) of Channa species.
Results and discussion
A total of 918 bp of fragments of 12S rRNA gene was sequenced for 75 individuals. All the sequences were aligned unambiguously with 735 bp in sequences of other Channa species in NCBI database. Due to its relative conservation in DNA sequence, 12S rRNA was used for the analysis of molecular evolution and phylogenetic studies (Skinner et al. Citation2013). Meanwhile, the abundance of polymorphism in mtDNA was showed in this study, the variation sites were 176; the parsimony informative polymorphic sites were 79 and the transition/transversion ratio up to 2.33, which indicated that the level of nucleotide substitution was unsaturated. These characteristics suggested that the mtDNA gene could be used as an effective molecular marker to identify different Channa species. 7 Haplotypes (WtcaHS1–S7) of white-type C. argus and 2 haplotypes (BtcaHS1–S2) of biocolour type C. argus was revealed by 12S rRNA gene (). The nucleotide sequences of all the tested Channa species and haplotypes were submitted in GenBank (KT358948–KT358951; KU852423–KU852433). Among the identified haplotypes, the sequences of WtcaHS1 and BtcaHS1, as well as the WtcaHB2 and BtcaHB1 were the common haplotypes with largest number of distribution.
Table 2. Haplotype information of 12S rRNA among white and biocolour type C. argus.
Taking Lateolabrax maculates and Epinephelus coioides as out-group, sequences alignment was performed via MEGA6.0. The results showed that the interspecific genetic distance ranged from 0.000 to 0.130 among 10 kinds of Channa species (). Interspecific sequence difference is commonly considered as a prerequisite for accurate identification of species for different species of generic and species level (Peng et al. Citation2009). Research shows that the genetic distances within species are generally less than 0.020, and most are less than 0.010 in 11 phylums 13,320 species of animals (Hebert et al. Citation2003). In this study, the intraspecific average genetic distances were 0.000/0.006 by analyzing the haplotype sequences, and the maximum genetic distances between different haplotypes were significantly less than 0.010. The genetic distances among other Channa species ranged from 0.023 to 0.130 except for C. maculata and C. maculata × C. argus. These results indicated that the white and biocolour-type C. argus have not yet reached the level of subspecies differentiation, which might correct the previous classification for the white type C. argus (Shih Citation1936). There is a similar report in the previous study, where Cynoglossus lighti and C. joyneri are considered as the same species in different morphological types since synonym phenomenon is an extremely common in the subfamily species of Cynoglossidae; the genetic distances between two species is merely 0.004 (Liu et al. Citation2010).
Table 3. Pairwise distance calculated using Kimura 2-parameter model for 12S rRNA.
As a gene sequence information, phylogenetic information can estimate the biological related group, genetic relationship, or effective and reflect the size of the development information of a gene, and estimate the accuracy by constructing a phylogenetic tree (Takashima et al. Citation2004). Molecular phylogenetic tree was constructed by the NJ method based on 12S rRNA gene using the Kimura 2-parameter model (). According to the comparative analysis of the construction of NJ tree, we found that the molecular phylogenetic tree was basically consistent, which showed that all haplotypes were clustered into a single group with a high confidence value between the white and biocolour type C. argus. From the phylogenetic tree constructed by haplotype sequences of the white and biocolour type C. argus, all haplotypes were clustered into one and cross each other. According to these findings, it could be concluded that the white-type C. argus should derive from biocolour type C. argus as an albino. This conclusion complements with the result obtained in our previous study where two molecular markers (16S rRNA) were performed in phylogenetic and germplasm analyses (Zhou et al. Citation2016). In order to further develop the classification in C. argus, more molecular markers will be performed in comprehensive analysis for further validation of the results in this study.
Acknowledgements
This work was supported by the Science and Technology Planning Project of Guangdong Province (2017A020225035); Produce-learn-research Project of Guangdong Province (2011B090400270); Fund Fostering Talents for Young Scholars of South China Agricultural University (201707N025); Talent introduction special funds of South China Agricultural University and Scientific Research Staring Foundation for Young Scholars of College of Marine Sciences. We also wish to express our appreciation to our anonymous reviewers for providing valuable comments on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Berra TM. 2007. Freshwater fish distribution. Chicago: University of Chicago Press.
- Bhat AA, Haniffa MA, Milton MJ, Paray BA, Divya PR, A G. 2014. Genetic variation of striped snakehead (Channa striatus Bloch, 1793) populations using random amplified polymorphic DNA (RAPD) markers. Int J Biodivers Conserv. 6:363–372.
- Cheng QT, Zheng BS. 1987. Systematic search of fish in China. 1st edition. Beijing: Science Press. p 456.
- Courtenay WR, Williams JD. 2004. Snakeheads (Pisces, Channidae): a biological synopsis and risk assessment. Denver, CO: US Geological Survey.
- Ding RH. 1994. The fishes of Sichuan, China. 1st edition. Chengdu: Sichuan Science and Technology Press.
- Hebert PDN, Cywinska A, Ball SL. 2003. Biological identifications through DNA barcodes. Proc Roy Soc Lond B. 270:313–321.
- Kimura S. 1934. Description of the fishes collected from the Yangtze-kiang, China, by the late Dr. K. Kishinouye and his party in 1927–1929. J Shanghai Sci Inst. 3:11–247.
- Liu SF, Liu JX, Zhuang ZM, Gao TX, Han ZQ, Chen DG. 2010. Monophyletic origin and synonymic phenomena in the sub-family Cynoglossinae inferred from mitochondrial DNA sequences. Biodivers Sci. 18:275–282.
- Peng JL, Wang XZ, Wang D, He SP. 2009. Application of DNA barcoding based on the mitochondrial COI gene sequences in classification of culter (Pisces: Cyprinidae). Acta Hydrobiol Sin. 2:271–276.
- Shih HJ. 1936. Notes on the labyrinth fishes of China. Bull Fan Mem Lnst Biol Peiping (Zool). 7:81–82.
- Skinner A, Hutchinson MN, Lee MSY. 2013. Phylogeny and divergence times of Australian Sphenomorphus group skinks (Scincidae, Squamata). Mol Phylogenet Evol. 69:906–918.
- Su SQ, Xiong B. 2001. Experiments in the artificial propagation of white snakehead. Reservoir Fish. 19:19–20.
- Takashima S, Ise H, Zhao P, Akaike T, Nikaido T. 2004. Human amniotic epithelial cells possess hepatocyte-like characteristics and functions. Cell structure and function. 29:73–84.
- Wang JX, Zhao XF, Zhuo CW, Liao ZG. 1992. Comparative studies on the ophiocephalus argus and O. A. Kimurai, With the systematics of Ophiocephalus argus. Trans Oceanol Limnol. 2:51–57.
- Wang JX, Liao ZG, Zhang ZG, Zhao XF. 1993. Principal components analysis of the Ophicephalus argus complex (Pisces: Channidae). J Southwest China Normal Univ (Natural Science). 18:168–172.
- Zhou AG, Zhuo XL, Zou Q, Chen JT, Zou JX. 2015. Population genetic diversity of the northern snakehead (Channa argus) in China based on the mitochondrial DNA control region and adjacent regions sequences. Mitochondrial DNA. 3:341–349.
- Zhou AG, Wang C, Jiang WZ, Li ZG, Chen YF, Xie SL, Luo JZ, Zou JX. 2016. Genetic comparison of two color morphs of northern snakehead (Channa argus) and Genetic relatedness among the family Chnannidae. Mitochondrial DNA Part A. 6:1–3.
