Abstract
In this study a Sipuncula species Phascolosoma sp. was collected from seagrass area from Chuuk lagoon Micronesia and its complete mitochondrial genome analyzed. This is the second complete mitochondrial genome record from the genus after Phascolosoma esculenta. The total length of mitochondrial genome of the species is 16,571 bp, which is longer than P. esculenta record. Also, locations of tRNA-Gly and putative control region are different between two records. Furthermore, phylogenetic relationship of Phascolosoma sp. are investigated due to protein-coding genes of mitochondrial genome. Due to the lack of recorded data, P. esculenta has been observed is the closest species to Phascolosoma sp. and they are belonging to the monophyletic group.
The genus Phascolosoma is one of the most species-rich genera in the phylum Sipuncula (Saiz et al. Citation2014) with the 25 valid species (WoRMS Editorial Board Citation2017). Almost all of these species are found in shallow water of the world’s oceans and inhabit cavities in solid substrates (Schulze et al. Citation2005). Although the many species are available, there is only one mitochondrial genome recorded from the genus which is Phascolosoma esculenta collected from China (Shen et al. Citation2009). In this study, a new complete mitochondrial genome from the genus Phscolosoma sp. is reported.
Specimens of Phscolosoma sp. have been collected from the sea grass area of Weno Island, Chook Lagoon, Federated States of Micronesia (7°26′41.0″N 151°53′57.7″E) on February 2015. The specimen genus has been identified morphologically and the collected specimen has been deposited in the Department of Life Science, Sangmyung University, with accession number SMU000510. Mitochondrial DNA sequencing, analysing, and phylogeny reconstruction methods were described in our previous study (Karagozlu et al. Citation2016).
Mitochondrial genome size of Phscolosoma sp. is evaluated as 16,571 bp long (GenBank accession number KX814447) and it is composed of 13 protein-coding genes, 2 ribosomal RNA genes, and 23 tRNA genes with base composition of 27.6% A, 22.4% C, 15.5% G, and 33.5% T. There are two overlapping regions in the genome and both are 1 bp lengths. Also, 31 intergenic sequences showed length variation ranging from 1 to 1116 bp. The gene order of the genome shows some difference from to that of P. esculenta. The main differences between structures of complete mitochondrial genomes are locations of tRNA-Gly and putative control region. The putative control region is located between tRNA-Glu and tRNA-Val with 459 bp length in the mitogenome of Phascolosoma sp. The same region is located between tRNA-Leu and NAD1 genes with 585 bp length in the mitogenome of P. esculenta. On the other hand, tRNA-Gly is located between ATP8 and tRNA-Gln genes in the mitogenome of Phscolosoma sp. while it located between tRNA-Val and ATP8 genes in the P. esculenta record. Also, there is tRNA-Arg in the mitogenome of Phscolosoma sp. which located between ATP6 and tRNA-His genes. This gene was not record in the P. esculenta mitogenome.
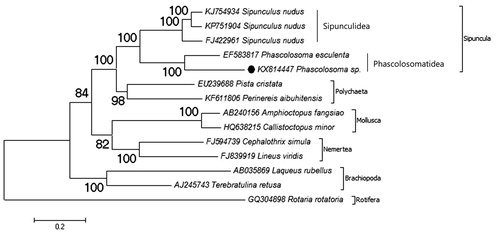
Phylogenetic relationship of Phscolosoma sp. in Trochozoa is investigated () in this study. The Trochozoa are composed of Sipuncula, Polychaeta, Mollusca, Annelida, Nemertea, and Brachiopoda phyla (Hejnol Citation2010). Due to reconstructed phylogenetic tree, P. esculenta is the closest species to Phscolosoma sp. and they belong to monophyletic group. The monophyly of the genus was previously well supported by nuclear and mitochondrial-based study (Kawauchi et al. Citation2012). The phylogenetic relationships of the Trochozoans due to mitochondrial genome-based phylogenetic study were supported by several molecular phylogenetic studies (Hausdorf et al. Citation2007; Witek et al. Citation2009; Hausdorf et al. Citation2010). Besides, there is a sister group relationship between Sipunculidea and Phascolosomatidea classes. The data show the similarity with previous transcriptomic-based study (Lemer et al. Citation2015). This genome data will be a part of mitochondrial genome library to provide evolutionary and systematic studies for the phylum Sipuncula.
Figure 1. Phylogenetic relationship of Phascolosoma sp. (KX814447) in the Trochozoa. For reconstruction of the phylogenetic tree, two species from the every Trochozoa phylum selected and a Rotifera species selected as the out group. The mitochondrial genome of the selected species retrieved from the GenBank. The presented record marked with a dot.
Acknowledgements
This research was a part of the project titled ‘Development of overseas marine bioresources and a system for their utilization’, funded by the Ministry of Oceans and Fisheries, Korea (Grant PM 59120). We thank to the Department of Marine Resources, State of Chuuk, Federated States of Micronesia, for allowing marine organism research.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Hausdorf B, Helmkampf M, Meyer A, Witek A, Herlyn H, Bruchhaus I, Hankeln T, Struck TH, Lieb B. 2007. Spiralian phylogenomics supports the resurrection of Bryozoa comprising Ectoprocta and Entoprocta. Mol Biol Evol. 24:2723–2729.
- Hausdorf B, Helmkampf M, Nesnidal MP, Bruchhaus I. 2010. Phylogenetic relationships within the lophophorate lineages (Ectoprocta, Brachiopoda and Phoronida). Mol Biol Evol. 55:1121–1127.
- Hejnol A. 2010. A twist in time-the evolution of spiral cleavage in the light of animal phylogeny. Integrat Compar Biol. 50:695–706.
- Karagozlu MZ, Sung JM, Lee JH, Kwon T, Kim CB. 2016. Complete mitochondrial genome sequences and phylogenetic relationship of Elysia ornata (Swainson, 1840) (Mollusca, Gastropoda, Heterobranchia, Sacoglossa). Mitochondrial DNA Part B. 1:230–232.
- Kawauchi GY, Sharma PP, Giribet G. 2012. Sipunculan phylogeny based on six genes, with a new classification and the descriptions of two new families. Zool Scripta. 41:186–210.
- Lemer S, Kawauchi GY, Andrade SCS, Gonzalez VL, Boyle MJ, Giribet G. 2015. Re-evaluating the phylogeny of Sipuncula through transcriptomics. Mol Phylogenet Evol. 83:174–183.
- Saiz JI, Cartes J, Mamouridis V, Mechó A, Pancucci-Papadopoulou MA. 2014. New records of Phascolosoma turnerae (Sipuncula: Phascolosomatidae) from the Balearic Basin, Mediterranean Sea. Marine Biodiversity Records. 7:e16.
- Schulze A, Cutler EB, Giribet G. 2005. Reconstructing the phylogeny of the Sipuncula. Hydrobiologia. 535:277–296.
- Shen X, Ma X, Ren J, Zhao F. 2009. A close phylogenetic relationship between Sipuncula and Annelida evidenced from the complete mitochondrial genome sequence of Phascolosoma esculenta. BMC Genomics. 10:136.
- Witek A, Herlyn H, Ebersberger I, Mark Welch DB, Hankeln T. 2009. Support for the monophyletic origin of Gnathifera from phylogenomics. Mol Phylogenet Evol. 53:1037–1041.
- WoRMS Editorial Board. 2017. World register of marine species [Internet]; [cited 2017-03-05]. Available from http://www.marinespecies.org at VLIZ.
