Abstract
The complete mitochondrial genome of the Asian warty newt Paramesotriton chinensis was sequenced. The complete mitogenome of P. chinensis is a circular double-stranded DNA sequence that is 16,361 bp long and was biased toward A + T content at 61.3% (33.0% A, 28.3% T, 23.9% C, and 14.7% G). The complete mitogenome of P. chinensis consists of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 1 ribosomal RNAs (16S rRNA), and 1 putative control region. This study presented the complete mitogenome of P. chinensis and provided essential and important DNA molecular data for further phylogenetic and evolutionary analysis for genus Paramesotriton.
The salamandrid genus Paramesotriton, currently consists of 14 nominal species endemic to southern China, except for P. deloustali which is mainly distributed in northern Vietnam. This genus commonly known as the Asian warty newts, consists of two species groups, which have been recovered as reciprocally monophyletic sister groups in molecular phylogenetic analyses (Gu et al. Citation2012; Yuan et al. Citation2014, Citation2016). Recent studies indicate several investigators who established different phylogenetic relationships for Paramesotriton based on different molecular markers (Weisrock et al. Citation2006; Gu et al. Citation2012; Yuan et al. Citation2014, Citation2016). The advent of molecular studies and the increasing number of genes and DNA regions used in the phylogenetic studies led to structuring of the taxonomy of Paramesotriton, improved understanding of the phylogenetic relationships of species. Paramesotriton chinensis is distributed in Zhejiang and Jiangxi Province, China. In this study, we propose to determine the complete mitochondrial genome using next-generation sequencing (NGS), which may verify the phylogenetic position of this species.
Specimen of P. chinensis was obtained from Zhejiang and deposited at the Zhejiang Museum of Natural History, Hangzhou, Zhejiang, China. Genomic DNA extraction and next-generation sequencing were described in previous publication (Shen et al. Citation2016). Initially, the raw next generation sequencing reads generated from HiSeq 2000 (Illumina, San Diego, CA) were altered to remove low-quality reads. Around 0.06% raw reads (7944 out of 13,849,558) were subjected to de novo assembly using commercial software (Geneious V9, Auckland, New Zealand) to produce a single, circular form of complete mitogenome with about an average of 122 × coverage. The mitogenomic sequence has been deposited into GenBank under the accession number KY609177.
The complete mitogenome of P. chinensis is a circular double-stranded DNA sequence that is 16,361 bp long and was biased toward A + T content at 61.3% (33.0% A, 28.3% T, 23.9% C, and 14.7% G). DOGMA (Wyman et al. Citation2004), ARWEN (Laslett & Canback Citation2008), and MITOS (Bernt et al. Citation2013) programs were used to predict the protein-coding, rRNA and tRNA genes of P. chinensis mitogenome. The complete mitochondrial genome of P. chinensis had the same gene arrangement and similar compositions according with other Paramesotriton species.
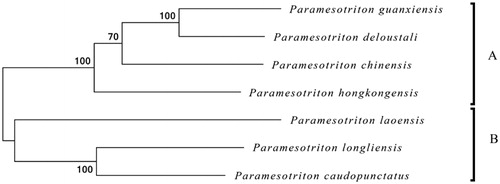
To further explore its taxonomic status and the phylogenetic relationship within Paramesotriton, we used MEGA6 software (Tamura et al. Citation2013) to construct a Maximum likelihood tree (with 500 bootstrap replicates) containing complete mitogenomes of seven species. These seven Paramesotriton species are recovered as a monophyletic group strongly supported, and could be divided into clades A (P. hongkongensis, P. chinensis, P. deloustali, and P. guangxiensis) and clade B (P. caudopunctatus, P. laoensis and P. longliensis) (). Our molecular data also reveal a close phylogenetic relationship among P. chinensis, P. deloustali, and P. guangxiensis, and this result was supported by previous phylogenetic studies (Gu et al. Citation2012; Yuan et al. Citation2014). This study presented the complete mitogenome of P. chinensis and provided essential and important DNA molecular data for further phylogenetic and evolutionary analysis for genus Paramesotriton.
Figure 1. Maximum likelihood (ML) phylogenetic tree based on the complete mitochondrial genome sequences of 6 closely related Paramesotriton species Numbers at the branches indicate bootstrapping values with 500 replications. Accession number: Paramesotriton longliensis (NC032310), Paramesotriton caudopunctatus (EU880326), Paramesotriton laoensis (EU880328), Paramesotriton guanxiensis (NC032309), Paramesotriton deloustali (EU880327), Paramesotriton hongkongensis (NC006407).
Acknowledgements
We thank Bihao Ji and Liangwei Sun from Zhejiang Dayanghu National Nature Reserve Administration for their field assistance. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Gu X, Chen R, Tian Y, Li S, Ran J. 2012. A new species of Paramesotriton (Caudata: Salamandridae) from Guizhou Province, China. Zootaxa. 3510:41–52.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Shen KN, Yen TC, Chen CH, Li HY, Chen PL, Hsiao CD. 2016. Next generation sequencing yields the complete mitochondrial genome of the lathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA. 27:1758–1759.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Weisrock DW, Papenfuss TJ, Macey JR, Litvinchuk SN, Polymeni R, Ugurtas IH, Zhao E, Jowkar H, Larson A. 2006. A molecular assessment of phylogenetic relationships and lineage accumulation rates within the family Salamandridae (Amphibia, Caudata). Mol Phylogenet Evol. 41:368–383.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yuan Z, Zhao H, Jiang K, Hou M, He L, Murphy RW, Che J. 2014. Phylogenetic relationships of the genus Paramesotriton (Caudata: Salamandridae) with the description of a new species from Qixiling nature reserve, Jiangxi, southeastern China and a key to the species. Asian Herpetol Res. 5:67–79.
- Yuan Z, Wu Y, Zhou J, Che J. 2016. A new species of the genus Paramesotriton (Caudata: Salamandridae) from Fujian, southeastern China. Zootaxa. 4205:549–563.
