Abstract
Phoxinus lagowskii is a freshwater fish that is widely distributed in China. In this study, a comparative analysis of the mtDNA control region (D-loop) was performed to analyze the natural population structure and genetic diversity of 54 individuals from eleven locations (T1, T2, T3, T4, T5, T6, T7, T8, T9, T10 and T11) which was divided with reservoirs. The estimated haplotype and nucleotide diversity were 0.734 and 0.03514, respectively. An AMOVA indicated that 79.78% of the total variation originated from individual populations and 20.22% came from variation within the 11 geographic populations, which showed high genetic differentiation among the 11 geographic groups. A test of neutral evolution and mismatch distribution indicated that historical expansion occurred in these populations. However, the findings of low genetic diversity and high genetic differentiation demonstrated that the reproduction isolated by reservoir has showed a certain effect for the development of the populations, and the results should provide new information for the conservation and exploitation of this species.
Introduction
The Amur minnow (Phoxinus lagowskii), a member of the family Cyprinidae, subfamily Leuciscus, is a common edible fish that is abundant and widely distributed in the Yangtze River, Qingjiang River, Hanjiang River and northern Yellow River in China (Guo & Wu Citation2012; Wang et al. Citation2013). Additionally, as the dominant species, the Amur minnow is widely distributed in the Liaohe River, the sampling site of the present study. A segment of the migratory route of the Amur minnow was sealed off by a glacier during the last glaciations, isolating a subpopulation of this species; thus, the Amur minnow is not contiguously distributed in streams, leading to its long-term isolation in the reservoir (Wang et al. Citation2005; Ye et al. Citation2005). Thus, Phoxinus lagowskii is an ideal model organism for the study of the geographic formation of the Liaohe River.
In recent years, due to a variety of human-caused threats and environmental pollution, the number of Amur minnow has decreased rapidly (Zhang Citation2006). Because of their very limited numbers and their high sensitivity to environmental factors, in 1988, this species was listed as a second-class state-protected wild animal in the China Red Data Book of Endangered Animals (Yue & Chen Citation1998). However, information on this species remains very scarce, and it is urgent for researchers to develop methods to protect this prized fish resource as soon as possible (Zhao & Zhang Citation2009; Kang, et al. Citation2011; Si et al. Citation2012). And with the completion of several reservoirs in the Liaohe River before 70s of the last century (Dong & Shi Citation1989), production isolation should be take place on the populations of Phoxinus lagowskii. How serious is the risk for the genetic diversity of Phoxinus lagowskii? The study of the population dynamics of Phoxinus lagowskii is important for the conservation of its endemic germplasm resources and genetic diversity.
As an effective molecular marker due to its rapid rate of evolution and maternal inheritance (Sun et al. Citation2012), mitochondrial DNA (mtDNA) has been widely applied to the study of population genetic structure, the origin and evolution of species, and genetic relationships(Weiss et al. 2011; Shi et al. Citation2012; Liu et al. Citation2015). In the present study, the mitochondrial control region (D-loop) was used to analyze the population genetic diversity and adaptive variation of the Phoxinus lagowskii from 11 populations in Liaoning Province. The results will be useful for the management of the germplasm resource, biodiversity conservation and exploitation and for better understanding the effects of habitat fragmentation on the genetic structure of the species to lay the foundation for effective protection and management strategies.
Materials and methods
Materials
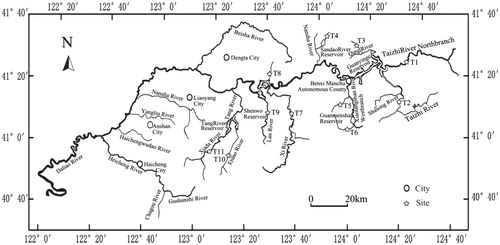
Adult Phoxinus lagowskii were obtained in December from 11 different sampling points that at least were separated by a reservoir in the Liaohe River (), Liaoning Province, China. Fin clips were collected and stored in 95% ethanol until DNA extraction.
Extraction of genomic DNA
Total genomic DNA was extracted from the fin tissue samples by a modified phenol/chloroform extraction method (Sambrook et al. Citation1989): the ethanol-preserved fin tissues (0.1 g) were dried for 30 min at 70 °C and then washed twice with ddH2O, followed by digestion in 600 ml of tissue extract (50 mM Tris–HCl (pH 8.0), 100 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 1% SDS) and 5–8 ml 20 mg/ml proteinase K for 4 h at 50 °C with constant gentle mixing. After centrifugation at 12,000 rpm for 10 min, the supernatant (DNA extract) was collected in a new 1.5-ml microcentrifuge tube and was sequentially extracted with 300 ml each of phenol and chloroform. The total DNA was recovered by overnight precipitation in 1 ml volumes of frozen absolute ethanol and washed twice by 75% ethanol at 12,000 rpm. The total DNA was standardized to a concentration of 50 ng/ml and stored in ddH2O.
Amplification of D-loop sequences
The mitochondrial D-loop was amplified by polymerase chain reaction (PCR) using the forward primer DL1: 5'-caa cat gcc ggg cgt tca tg-3' and the reverse primer DL2: 5'-gcg atg gct aac cgt agc tc-3'. Amplifications were carried out in 30 μl reaction volumes containing the following components: 4 μl of genomic DNA, 23.2 μl of golden mix (Golden Easy PCR System; TaKaRa, Dalian, China), 1.2 μl of each primer, 0.4 μl of Taq polymerase (TaKaRa, Dalian, China). The PCR conditions were as follows: 95 °C for 3 min followed by 35 cycles of denaturing at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, and a final extension at 72 °C for 10 min. All of the amplified products were purified with a Gel Extraction Kit (Generay Biotech (Shanghai) Co., Ltd., Shanghai, China) following the manufacturer’s instructions. The PCR products were sent to Shanghai Generay Biotech Co. Ltd. (Shanghai, China) for sequencing.
Sequence alignments and analyses
The sequences were proofread, edited and aligned with BioEdit version 7.0.5.3 (Ibis Biosciences, Carlsbad, CA). The polymorphism was assessed using DnaSP version 5.0 (Universitat de Barcelona, Barcelona, Spain). Arlequin software version 3.11 (University of Berne, Berne, Switzerland) was used to estimate the genetic differentiation and Fst values with an analysis of molecular variance (AMOVA) model (Excoffier et al. Citation2005). The historical population expansion was examined using Tajima’s (Citation1989) and Fu’s (Fu Citation1997) statistics with 10,000 demographic history records among the four populations. The parameters of the genetic diversity within and between populations, such as the haplotype diversity (Hd), nucleotide diversity (Pi), average number of nucleotide difference (K) and gene flow (Nm), were estimated with MEGA software (version 5.05), and the phylogenetic reconstruction of the haplotypes was also conducted using the neighbour-joining (NJ) method performed using MEGA version 5.05 from a bootstrap consensus of 10,000 replicates (Kumar et al. Citation2004; Tammura et al. Citation2007).
Results
Composition analysis of the D-loop sequence
Approximately 300 bp of the mtDNA control region of 11 populations was obtained (), which shared high homology with other cyprinid fishes. A total of 54 sequences were analyzed, and the estimated transition/transversion bias (R) was 5.00, A = 29.6, C = 27.8, G = 14.9 and T = 27.8, with (A + T) (57.4%) significantly higher than (C + G) (42.6%) ().
Table 1. Base composition of the mitochondrial D-loop gene sequence of eleven populations of Phoxinus lagowskii.
Analysis of genetic diversity and population genetic structure
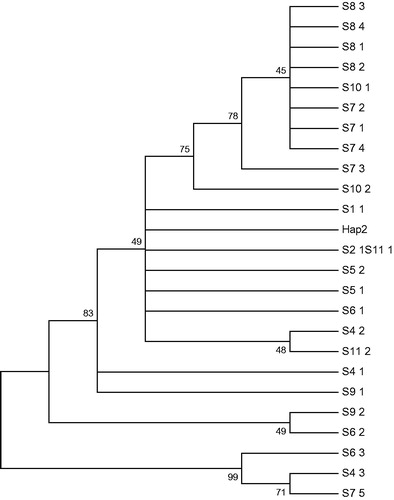
Twenty-five D-loop haplotypes from 54 individuals (Phoxinus lagowskii) () were obtained using DnaSP software version 5.0. Haplotype Hap_2 was shared among nine populations, Haplotype Hap_1 was separately observed in the T2 and T11 populations, and the other haplotypes were unique to each population. Five haplotypes were observed in the T7 population, which is the most in any of the populations, and only one haplotype was observed in the T3 population, which is the least.
Table 2. Distribution of the 25 haplotypes in Phoxinus lagowskii populations.
The estimates of haplotype diversity and nucleotide diversity are shown in . The T7, T8 and T9 populations showed the highest haplotype diversity (h = 1.000), followed by the T5 population (h = 0.833), the T4 and T6 populations (h = 0.8), the T10 and T2 populations (h = 0.714; h = 0.7, respectively), with the T3 population exhibiting the lowest haplotype diversity. The total haplotype diversity of the T11 population was 0.734. The T7 population exhibited the highest level of nucleotide diversity (π = 0.08142) among the 11 populations.
Table 3. Haplotype diversity (h) and nucleotide diversity (π) of Phoxinus lagowskii populations.
Analysis of molecular variance
The AMOVA model (Meirmans Citation2006) was used to test the significance of the 11 population structures based on the haplotype frequency; 20.22% of the molecular variance was attributed to the differentiation among populations, whereas 79.78% of the molecular variance was observed within populations ().
Table 4. Analysis of molecular variance (AMOVA) for the Phoxinus lagowskii populations.
The tests of neutral evolution (Tajima’s D and Fu’s Fs) indicated a positive value (in population T8), but there was no significant difference (p > .05) among the 11 populations; the topology of the resulting neighbour-joining (NJ) tree () revealed that population T8 is relatively closed, and the other populations are dispersed. The above analysis indicated no significant population expansion in the 11 populations.
Discussion
Analysis of the structure of the mtDNA control regions of Phoxinus lagowskii
Phoxinus lagowskii is mainly distributed in the Heilongjiang River, Tumen River, Liaohe River, Yellow River and the middle reaches of the Yangtze River in China. Phoxinus lagowskii is the dominant carp species in the Liaohe River of Liaoning Province (Zheng & Zhang Citation2002).
The MtDNA control region sequences of 54 individuals were investigated and analyzed in this study. The D-loop region sequence is located between the tRNA-Pro and tRNA-Phe genes. The conserved sequence domain structure of the control regions showed high similarity with that reported for other Cyprinidae, i.e. Phoxinus oxycephalus, Rhynchocypris lagowskii, Rhynchocypris oxycephalus and Phoxinus steindachneri (Wang et al. Citation2011) The nucleotide microsatellite has also been found in some mammals and teleosteans (Yue et al. Citation2006); for example, Cyprinid fish possessed an AT-type microsatellite in the mtDNA D-loop. The differences in the nucleotide microsatellite can be used for the identification of separate families of fishes.
Analysis of the genetic diversity of Phoxinus lagowskii
Twenty-five haplotypes were obtained based on the 54 individuals. The major haplotype (Hap_2), as a common haplotype among the nine populations, supports the fact that these populations are descended from a common ancestor, whereas the other twenty-four haplotypes separately belong to different populations. All twenty-five haplotypes were associated with the dispersion and expansion of the population (). Therefore, the eleven populations appear to have diverged from each other in recent evolution. The cluster analysis supported the view that the fish were able to pass through the reservoir in the artificial drainage due to their smaller body size and subsequently mixed with populations from other regions, enabling gene flow, which explains the haplotype cross-phenomenon. We could infer this from the situations of the 11 populations that the population has experienced growth, reproduction and population growth in recent years. The result is consistent with the hypothesis that the subspecies has undergone a long isolation and evolutionary bottleneck (Miracle & Campton Citation1995). The results of the evolutionary analysis not only exhibited some intraspecific differences but also showed divergent and adaptive variance resulting from the long isolation of the different geographic populations and suggested that eight populations have experienced a long isolation and genetic divergence compared to other populations.
The analysis of the genetic structure of the eleven populations revealed that haplotype and nucleotide diversities were an important index of population genetic diversity. The haplotype and nucleotide diversities of the 54 individuals collected from the eleven sampling points were 0.7340 and 0.03514, respectively. The T7, T8 and T9 populations exhibited the highest haplotype diversity (h = 1.000), whereas the T7 population exhibited the highest nucleotide diversity (π = 0.08142). The Phoxinus lagowskii in the Liaoning River exhibit high genetic diversity. Nevertheless, this rare fish resource should be protected from the degeneration of biodiversity.
In recent years, populations of Phoxinus lagowskii have suffered severe population declines; in the past 40 years, some populations have totally disappeared due to increases in the local water temperature and excessive fishing by humans (Liang Citation2009).The distribution range of this species is shrinking. National and local natural reserves (Liaohe River Natural Reserve in Panjin) have been established by local governments to protect the ecosystems and biodiversity. The population of the Phoxinus lagowskii in the Liaohe River has adapted to changing environmental and demographic events (Sun et al. Citation2016). In recent years, many water drawdown projects have been implemented on the Liaohe River; although these projects have had no impact on fish populations in the short term, they will lead to the reduction of biodiversity of endangered species and even extinction in the long term. Therefore, it is important to consider gene exchange when water conservancy projects are constructed to ensure the sustainable development of the genetic resources of the biological population.
The differentiation of haplotypes in Phoxinus lagowskii conformed with the results of the AMOVA revealing that variation within the population accounted for 79.78% of the total variation and variation among populations was only 20.22%; therefore, genetic variation does not occur primarily in individual populations. Thus, it was concluded that the genetic differentiation among the eleven populations was low. Despite the low level of genetic differentiation, the results of the present study also support substantial differences in geographic structure associated with different population drainages. As above, the results of this research demonstrated that the reproduction isolated by reservoir has showed a certain effect for the development of Phoxinus lagowskii populations, and the results should provide some new information to the government and scientific researchers for the conservation and exploitation of this species.
Acknowledgements
We would like to express our gratitude to Professor Jiang Zhiqiang and the anonymous reviewers for their critical comments on the manuscript. This work was jointly supported by grants from the National Natural Science Foundation of China (41101481), National High-Tech R&D Program (863 Program) (2012AA10A413-3).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dong SL, Shi WL. 1989. The plankton of the reservoir of Liaoning. J Dalian Fish Univ. 1989:1–10.
- Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform. 1:47–50.
- Fu YX. 1997. Statistical test of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 147:915–925.
- Guo GL, Wu Y. 2012. Introduction of new varieties of aquaculture: Phoxinus lagowskii Dybowski. Guide Fish. 18:38–39. (in Chinese)
- Kang X, Zhang Y, Zhang N, Ding S, Meng W, Li XD. 2011. Assessment of Habitat suitability of juvenile Phoxinus lagowskii Dybowski in Taizi River. Asia J Ecotoxicol. 6:310–320.
- Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformat. 5:150–163.
- Liang TH, Xie XM, Cui XY, Yang LL. 2009. Study on water resources rational allocation in West Liao River Basin. J Chin Ins Water Res Hydropower Res. 42:2214–2217.
- Liu HX, Li Y, Liu XL, Xiong DM, Wang LX, Zou GW, Wei QW. 2015. Phylogeographic structure of Brachymystax lenok tsinlingensis (Salmonidae) populations in the Qinling Mountains, Shaanxi, based on mtDNA control region. Mitochondrial DNA. 26:532–537.
- Meirmans PG. 2006. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 60:2399–2402.
- Miracle AL, Campton DE. 1995. Tandem repeat sequence variation and length heteroplasmy in the mitochondrial DNA D-loop of the threatened Gulf of Mexico sturgeon, Acipenser oxyrhynchus desotoi. J Hered. 86:22–27.
- Sambrook J, Fritsch EF, Maniatis T. 1989. T-molecular cloning. New York: Cold Spring Harbor Press.
- Shi W, Kong XY, Jiang JX, Miao XG. 2012. Preliminary study on the rapid evolution of mtDNA control region and the elongated mechanism of tandem repeat units in cynoglossinae fishes. Period Ocean Univ China. 42:81–87. (in Chinese)
- Si S, Wang Y, Xu GF, Yang S, Mou Z, Song Z. 2012. Complete mitochondrial genomes of two lenoks, Brachymystax lenok and Brachymystax lenok tsinlingensis. Mitochondrial DNA. 23:338–340.
- Sun P, Shi Z, Yin F, Peng S. 2012. Genetic variation analysis of Mugil cephalus in China sea based on mitochondrial COI gene sequences. Biochem Genet. 50:180–191.
- Sun Q, Wang D, Wei Q. 2016. The complete mitochondrial gemone of Phoxinus lagowskii (Teleostei, Cypriniformes: Cyprinidae). Mitochondrial DNA. 27:1–2.
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–595.
- Tammura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 8:1596–1599.
- Wang ML, Li BM, Jiang YS, Meng SY, Zhang YZ, Su X, Jiang ZQ. 2013. Study on individual fecundity of Phoxinus lagowskii Dybowski in Taizi River of Benxi city. J Yangtze Univ. 10:28–33. (in Chinese)
- Wang Y, Zhang X, Yang S, Song Z. 2011. The complete mitochondrial genome of the taimen, Hucho taimen, and its usual features in the control region. Mitochondrial DNA. 4:111–119.
- Wang ZS, Lu C, Xu CR, Lei GC. 2005. Impact of river-lake isolation on the spatial distribution pattern of Hemisalanx brachyrostralis. Biodiver Sci. 13:407–415. (in Chinese)
- Weiss S, Maric′ S, Snoj A. 2011. Regional structure despite limited mtDNA sequence diversity found in the endangered Huchen, Hucho hucho (Linnaeus, 1758). Hydrobiologia. 658:103–110.
- Ye SF, Ding DW, Wang WH. 2005. Large-scale estuarine engineering and estuarine habitat fragmentation of water body in the Yangtze River Estuary. Acta Ecologica Sinica. 25:265–272. (in Chinese)
- Yue GH, Liew WC, Orban L. 2006. The complete mitochondrial genome of a basal teleost, the Asian arowana (Scleropages formosus, Osteoglossidae). BMC Genom. 7:242.
- Yue PQ, Chen YY. 1998. China Red Data Book of Endangered Animals (Pisces). Beijing: Science Press. (in Chinese)
- Zhang ZW. 2006. Textbook of Biodiversity(K. V. Krishnamurthy book). New Hanover (NC): Science Publishers.
- Zhao YH, Zhang CG. 2009. Threatened fishes of the world: brachymystax lenok tsinlingensis Li, 1966 (Salmonidae). Environ Biol Fish. 86:11–12.
- Zheng BR, Zhang YP. 2002. Variation analysis of mtDNA control region sequences of the common carp. J Fish China. 26:289–294. (in Chinese)