Abstract
The mitochondrial genome of Rhinolophus yunnanensis (Chiroptera: Rhinolophidae) is a circular molecule of 16,865 bp in length with a base composition of 31.2% A, 24.3% T, 29.6% C, 14.9% G. In the control region of R. yunnanensis, the sequence of 5'-CAACGTATACACG-3′ repeats 18 times. Phylogenetic analyses indicate that R. yunnanensis is a sister clade to ((Rhinolophus sinicus sinicus + R. sinicus sinicus) + (R. macrotis + (R. pumilus + R. monoceros))).
Dobson’s horseshoe bats, Rhinolophus yunnanensis, are found in China, India, Myanmar and Thailand. Many species of Rhinolophus are extremely difficult to distinguish. Although nine complete mitochondrial genomes of Rhinolophus are available in GenBank, the complete mitochondrial genome of R. yunnanensis has not yet been reported. Hence, in this study, we sequenced the complete mitochondrial genome of R. yunnanensis to provide more molecular data to discuss the taxonomic analysis and phylogenetic relationship of Rhinolophus.
Samples of R. yunnanensis (No. GXGL20140711001–GXGL20140711004) were collected from Guangxi, China, and were identified by JY Zhang. The samples were deposited in the lab of Dr. JY Zhang, College of Life Sciences and Chemistry, Zhejiang Normal University. The total genomic DNA (No. GXGL20140711001) was extracted from liver tissues of R. yunnanensis using the Qiagen DNeasy Blood & Tissue Kit (50) (Hilden, Germany). The universal primers for PCR amplification were designed referring to previously published mitochondrial genomes of genera Myotis and Nyctalus (Kim et al. Citation2011; Nam et al. Citation2015; Yoon et al. Citation2015; Qian et al. Citation2016; Jiang et al. Citation2016; Wang et al. Citation2016; Yu et al. Citation2016) by primer premier 5.0 (PREMIER Biosoft International, Palo Alto, CA).
The complete mt genome of R. yunnanensis contains 22 transfer RNAs genes, 13 protein-coding genes, two ribosomal RNAs and non-coding regions, which is similar to the nine mitochondrial genomes from other Rhinolophus species (Nikaido et al. Citation2001; Lin et al. Citation2002; Xu et al. Citation2012; Yoon et al. Citation2013; Sun et al. Citation2016; Xie et al. Citation2016; Zhang et al. Citation2016; Xiao et al. Citation2017). The mt genome of R. yunnanensis is 16,865 bp in length. The total base composition of R. yunnanensis is 31.2% A, 24.3% T, 29.6% C, 14.9% G, with an A + T content of 55.5%. All protein-coding genes begin with ATN as the start codon. ND2, COIII and ND4 genes are terminated with an incomplete stop codon T, whereas ND1 ends with an incomplete stop codon TA, Cytb ends with AGA, and the other protein-coding genes end with TAA. The 35-bp sequence between the tRNAAsn and tRNACys genes is replication origin. The length of the control region of R. yunnanensis is 1420 bp with 54.9% AT content, and the sequence 5′-CAACGTATACACG-3′ repeats 18 times.
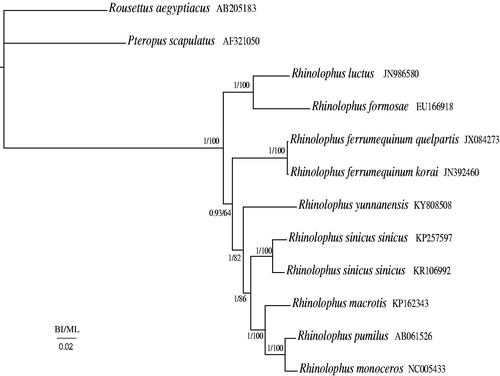
Phylogenetic relationships were reconstructed using the Bayesian inference (BI) method implemented in MrBayes version 3.1.2 (Huelsenbeck & Ronquist Citation2001) and maximum likelihood (ML) in RaxmlGUI 1.3 (Silvestro & Michalak, Citation2012) based on 13 protein-coding genes among 12 species including Rousettus aegyptiacus (AB205183 unpublished) and Pteropus scapulatus (Lin & Penny Citation2001) as outgroups. Phylogenetic relationships of BI and ML analyses produced highly concordant topologies (). In the BI and ML phylogenetic trees, R. yunnanensis is a sister clade to ((Rhinolophus sinicus sinicus + Rhinolophus sinicus sinicus) + (Rhinolophus macrotis + (Rhinolophus pumilus + Rhinolophus monoceros))) (1.00 in BI and 82% in ML), Rhinolophus luctus is a sister clade to Rhinolophus formosae (1.00 in BI and 100% in ML), and (Rhinolophus ferrumequinum quelpa + Rhinolophus ferrumequinum korai) is a sister clade to (R. yunnanensis + ((R. sinicus sinicus + R. sinicus sinicus) + (R. macrotis + (R. pumilus + R. monoceros)))) (0.93 in BI and 64% in ML).
Figure 1. Phylogenetic tree of the relationships among 12 species of Vespertilionidae, including R. yunnanensis based on the nucleotide dataset of the 13 mitochondrial protein-coding genes. The Bayesian posterior probability values and the maximum-likelihood bootstrap values are indicated above nodes. The GenBank numbers of all species are shown in the figure.
Nucleotide sequence accession number
The complete mitochondrial genome of R. yunnanensis has been assigned the GenBank accession number KY808508.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Jiang JJ, Wang SQ, Li YJ, Zhang W, Yin AG, Hu M. 2016. The complete mitochondrial genome of insect-eating brandt’s bat, Myotis brandtii (Myotis, Vespertilionidae). Mitochondrial DNA Part A. 27:1403–1404.
- Lin YH, McLenachan PA, Gore AR, Phillips MJ, Ota R, Hendy MD, Penny D. 2002. Four new mitochondrial genomes and the increased stability of evolutionary trees of mammals from improved taxon sampling. Mol Biol Evol. 19:2060–2070.
- Lin YH, Penny D. 2001. Implications for bat evolution from two new complete mitochondrial genomes. Mol Biol Evol. 18:684–688.
- Nam TW, Kim HR, Cho JY, Park YC. 2015. Complete mitochondrial genome of a large-footed bat, Myotis macrodactylus (Vespertilionidae). Mitochondrial DNA. 26:661–662.
- Nikaido M, Kawai K, Cao Y, Harada M, Tomita S, Okada N, Hasegawa M. 2001. Maximum likelihood analysis of the complete mitochondrial genomes of eutherians and a reevaluation of the phylogeny of bats and insectivores. J Mol Evol. 53:508–516.
- Kim YM, Choi EH, Kim SK, Jan KH, Ryu SH, Hwang UW. 2011. Complete mitochondrial genome of the Hodgson's bat Myotis formosus (Mammalia, Chiroptera, Vespertilionidae)). Mitochondrial DNA. 22:71–73.
- Qian KN, Yu DN, Cheng HY, Storey KB, Zhang JY. 2016. The complete mitochondrial genome of Nyctalus noctula (Chiroptera: Vespertilionidae). Mitochondrial DNA Part A. 27:2365–2366.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Sun HJ, Dong J, Shi HZ, Ren M, Hua PY. 2016. The complete mitochondrial genome of a Chinese rufous horseshoe bat subspecies, Rhinolophus sinicus sinicus (Chiroptera: Rhinolophidae). Mitochondrial DNA Part A. 27:3301–3302.
- Wang SQ, Li YJ, Yin AG, Zhang W, Jiang JJ, Wang WL, Hu M. 2016. The complete mitochondrial genome of David’s myotis, Myotis davidii (Myotis, Vespertilionidae). Mitochondrial DNA Part A. 27:1587–1588.
- Xiao YH, Sun KP, Feng J. 2017. Complete mitochondrial genomes of two subspecies (Rhinolophus ferrumequinum nippon and Rhinolophus ferrumequinum tragatus) of the greater horseshoe bat (Chiroptera: Rhinolophidae). Mitochondrial DNA Part A. 28:96–97.
- Xie L, Sun K, Feng J. 2016. Characterization of the complete mitochondrial genome of the Rhinolophus sinicus sinicus (Chiroptera: Rhinolophidae) from Central China. Mitochondrial DNA Part A. 27:2734–2735.
- Xu H, Yuan Y, He Q, Wu Q, Yan Q, Wang Q. 2012. Complete mitochondrial genome sequences of two Chiroptera species (Rhinolophus luctus and Hipposideros armiger). Mitochondrial DNA. 23:327–328.
- Yoon KB, Cho JY, Park YC. 2015. Complete mitochondrial genome of the Korean ikonnikov's bat Myotis ikonnikovi (Chiroptera: Vespertilionidae)). Mitochondrial DNA. 26:274.
- Yoon KB, Kim JY, Kim HR, Cho JY, Park YC. 2013. The complete mitochondrial DNA genome of a greater horseshoe bat subspecies, Rhinolophus ferrumequinum quelpartis (Chiroptera: Rhinolophidae). Mitochondrial DNA. 24:22–24.
- Yu DN, Qian KN, Storey KB, Hu YZ, Zhang JY. 2016. The complete mitochondrial genome of Myotis lucifugus (Chiroptera: Vespertilionidae). Mitochondrial DNA Part A. 27:2423–2424.
- Zhang L, Sun K, Feng J. 2016. Complete mitochondrial genome of the big-eared horseshoe bat Rhinolophus macrotis (Chiroptera, Rhinolophidae). Mitochondrial DNA Part A. 27:4078–4079.
