Abstract
Salmo trutta fario population is an important species for aquaculture and livestock industry. Moreover, these species were used for studies on the molecular markers. Mitochondrial genomic is also beneficial for phylogenetic studies in salmonid species. They applied maternal traits for mitogenome, whereas paternal traits related to nuclear genomics. It will be genetically highly divergent indicating that they may represent distinct and potentially locally adapted gene pools (Apostolidis et al., 2011). Evolutionary history has been studied on the Salmo taxa such as brown trout, Salmo salar, Salmo trutta populations (Kottelat & Freyhof, 2007; Simonovic et al., 2007).
Keywords:
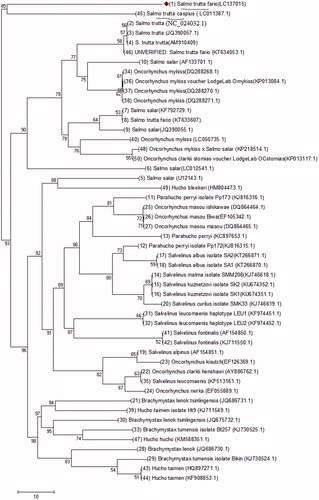
The evolutionary history of the Salmo taxa species including Salmo salar and Salmo trutta populations have been studied (Bernatchez Citation2001; Kottelat & Freyhof Citation2007). In the current study, 30 samples were collected from three major rivers in Iran: Cheshmeh Kileh, Ghasem Abad, and Jaj Roud. Total genomic DNA was isolated from muscle tissue (10 mg) taken from fresh specimens. The sample salmonids were caught in the Fall of 2015 and they were three years old. The left side was photographed and the right pectoral fin was clipped, tagged like the whole fish, and was subsequently fixed in 96% ethanol. DNA isolation was carried out following the phenol–chloroform protocol (Sambrook & Russel Citation2001). 10 ng of purified PCR product were sequenced. Mitogenomic study in S.t. fario is a new mitogenome sequenced for the first time in Iran and deposited in the GenBank (LC137015.1.) (). The evolutionary history was inferred using the neighbour-joining Method (Saitou & Nei Citation1987)). Evolutionary analyses were conducted using MEGA7 (Kumar et al. Citation2016). Here, the optimal tree with the sum of branch length equal to 638.75 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree, which were computed using the Maximum Composite Likelihood method (Tamura & Nei Citation1993). The analysis involved 50 nucleotide sequences. There were 16,622 positions in the final dataset. Results showed that a major diversity exists between S.t. fario and other salmonids like Hucho taimen, etc., S.t. fario and S.t. caspius are underlined in to show the close relationship observed between them.
Acknowledgments
This work was financially supported by the Research Council of Islamic Azad University, Tonekabon Branch, Iran.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernatchez L. 2001. The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution. 55:351–379.
- Kottelat M, Freyhof J. 2007. Handbook of European freshwater fishes. Cornol, Switzerland: Publications Kottelat.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY): CSH Laboratory Press.
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 4:406–425.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10:512–526.