Abstract
Three mitochondrial DNA regions were sequenced in two individuals of the Chukchi charr Salvelinus andriashevi from Lake Istihed (Chukotka): the entire gene sequences of cytochrome b and cytochrome c oxidase-1, and control region. The low level of sequence divergence detected between the sequences suggests that S. andriashevi is probably an isolated population of the Taranetz charr S. taranetzi. The genealogy of mtDNA haplotypes confirms the phylogenetic closeness of S. andriashevi, S. taranetzi, Salvelinus sp. 4 (Lake Nachikinskoe), S. krogiusae (Lake Dal’nee), S. boganidae and S. elgyticus (Lake Elgygytgyn), and S. a. erythrinus (NWT).
The divergence of phenotypically different and often geographically isolated forms is a significant problem for the taxonomy and phylogeny of charr of the genus Salvelinus. A theme that should be considered separately is isolated lacustrine charr populations known in many water bodies of Northeast Asia. Among such charrs is the Chukchi charr, an endemic narrow-range species represented by a single population in land-locked Lake Istihed (64°29' N/173°33' W) in the Chukotka Peninsula (Chereshnev et al. Citation2002). Originally described as a separate species, Salvelinus andriashevi Berg (Citation1948) was subsequently synonymized with Salvelinus alpinus malma var. andriashevi (Barsukov Citation1958) or regarded as a subspecies Salvelinus alpinus (Glubokovsky et al. Citation1979). Relying on the identity of karyotypes (Frolov & Frolova Citation2001) and 22 protein nuclear loci (Omel’chenko et al. Citation1998) from Istihed and Achchen lakes, it was concluded that the Chukchi charr represents an isolated local population of the Taranetz charr. However, the species status of Chukchi charr was later confirmed based on morphological characters (Chereshnev et al. Citation2002).
We have sequenced the three mitochondrial DNA (mtDNA) regions, among them those in S. andriashevi for the first time, for comparison with other lake charrs and for more precise phylogenetic analysis. Sequences of mtDNA were investigated in 70 specimens of charrs: S. andriashevi (Lake Istihed), S. taranetzi (Achchen and Pekulineiskoe Lakes, Chukotka), Salvelinus sp. 4 (Lake Nachikinskoe, Kamchatka), S. krogiusae (Lake Dal’nee, Kamchatka), S. boganidae and S. elgyticus (Lake Elgygytgyn, Chukotka), S. alpinus alpinus (Lake Sitasjaure, Scandinavian), S. alpinus oquassa (Floods Pond, North America), S. alpinus erythrinus (Jayko and Lauchlan Rivers, Victoria Island, Nunavut, Canada), S. malma malma (Paratunka and Kamchatka Rivers; Nachikinskoe, Achchen, and Pekulineiskoe Lakes). The fish specimens are stored in the collection of the Laboratory of Genetics (National Scientific Center of Marine Biology FEB RAS; www.imb.dvo.ru). Following the previous genetic studies with reference to Salvelinus species (Crête-Lafrenière et al. Citation2012; Yamamoto et al. Citation2014), the entire cytochrome b gene (Cytb; 1141 bp), a segment of the 5' end of the cytochrome c oxidase I gene (COI; 1200 bp) and control region (CR; 1021 bp) were sequenced using following primers (Ward et al. Citation2005; Uiblein et al. Citation2001). All nucleotide sequence data in this study are available from Genbank/NCBI under accession numbers KY1222045-KY122332. Two haplotypes were retrieved from Genbank to add to the analysis (accession numbers: S. malma malma KJ746618, S. alpinus AF154851).
The sequences are very similar in size and gene arrangement and composition to the charr genomes published previously (Doiron et al. Citation2002; Balakirev et al. Citation2016). The total length of mtDNA nucleotide sequences was 3362 bp. The overall base composition was 26.5% A, 29.8% T, 16.6% G and 27.2% C. The combined sequence (1-1021 bp СR, 1022-2221 bp COI, and 2222-3362 bp Cytb) had 124 variable sites (92 parsimony-informative) and 37 different haplotypes. The analyzed mtDNA region in two speciments of S. andriashevi (accession numbers: KY122239/KY122045/KY122142 and KY122240/KY122046/KY122143) had one haplotype. The haplotype of S. andriashevi is placed in a phylogenetic group combining haplotypes of S. taranetzi, Salvelinus sp. 4, S. krogiusae, S. elgyticus, S. boganidae and S. a. erythrinus (). The genealogical distance between this group and haplotypes of S. m. malma-S. alpinus was 42 nucleotide substitutions. S. andriashevi was most close to S. taranetzi from the type locality (Lake Achchen), the differences between the nearest haplotypes were two substitutions. The level of divergence between S. andriashevi and other taxa within the phylogenetic group was relatively low (Dxy = 0.001 ± 0.000 – 0.003 ± 0.001). These values were somewhat lower than those obtained based on the mtDNA PCR-RFLP analysis but corresponded to the level of intraspecific variability in the genus Salvelinus (Oleinik et al. Citation2015). At the same time, the level of divergence of allopatric S. andriashevi and S. malma malma (Dxy = 0.014 ± 0.002), S. andriashevi and S. alpinus alpinus (Dxy = 0.015 ± 0.002) matched the estimates for allopatric and sympatric populations of S. taranetzi and S. malma malma (Dxy = 0.014 ± 0.002), Salvelinus sp. 4 and S. malma malma (Dxy = 0.013 ± 0.002), S. krogiusae and S. malma malma (Dxy = 0.013 ± 0.002). Thus, the ratio of within- to between-population divergence of the mtDNA nucleotide sequences was analogous to that previously reported for S. taranetzi and S. m. malma populations (Oleinik et al. Citation2015).
Figure 1. Median-joining network for the charrs of the genus Salvelinus, resulting from the analysis of combined sequences of mtDNA (COI, Cytb genes, and CR). Mutational differences between the haplotypes are shown on the branches; the circle size is proportional to the absolute haplotype frequencies. All mutations have equal weights; the interval of the median vector search (black circles), ɛ is zero.
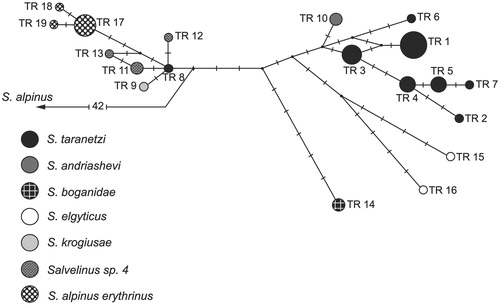
Our results suggest that the specimens of Chukchi charr belong to the arctic group of Taranetz charr according to Oleinik et al. (Citation2015). The genealogy of mtDNA haplotypes supports the phylogenetic closeness of S. andriashevi with S. taranetzi and their recent divergence and/or origin from a common ancestor. S. andriashevi is the least diverged in the arctic group and can be regarded as an isolated population of S. taranetzi. We encountered the problem of non-conformity of the taxonomic differentiation of charrs based on morphological and genetic analyses. Chereshnev et al. (Citation2002) argued for the independent status of Chukchi charr and emphasized that the degree of the craniological differences between S. andriashevi and S. taranetzi is comparable to the differences between S. andriashevi and S. malma and between S. andriashevi and S. alpinus. The objective reason for the observed discrepancy of morphological and genetic differentiation is likely to be recent divergence of the populations, and/or uneven evolutionary dynamics of qualitatively different characters against the background of exceptional ecological plasticity of charrs (Brunner et al. Citation2001; Oleinik et al. Citation2015). Another reason may be limitations imposed by some adaptive morphological characters as phylogenetic markers. Despite the low level of genetic divergence from S. taranetzi, the isolated charr population of Lake Istihed is of particular interest for the study of the problem of evolution and speciation in the Salvelinus group, as well as for the rational management and biological monitoring of Arctic ecosystems.
Disclosure statement
The authors declare no financial interest or benefit that has arisen from the direct applications of this research. The authors report no conflicts of interest, and are solely responsible for all content and writing.
Additional information
Funding
References
- Balakirev ES, Romanov NS, Ayala FJ. 2016. Complete mitochondrial genomes of the Northern (Salvelinus malma) and Southern (Salvelinus curilus) Dolly Varden chars (Salmoniformes, Salmonidae)). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1016–1017.
- Barsukov VV. 1958. Fishes of Provideniya Bay and adjacent waters of the Chukchi Peninsula. Proceedings of Zoological Institute, USSR Academy of Sciences. 25:131–163.
- Berg LS. 1948. Freshwater fishes of the USSR and adjacent countries, Vol 1. Moscow: USSR Academy of Sciences.
- Brunner PC, Douglas MR, Osinov A, Wilson CC, Bernatchez L. 2001. Holarctic phylogeography of arctic charr (Salvelinus alpinus L.) inferred from mitochondrial DNA sequences. Evolution. 55:573–586.
- Chereshnev IA, Volobuev VV, Shestakov AV, Frolov SV. 2002. Salmonoid fishes in Russian North-East. Vladivostok: Dalnauka.
- Crête-Lafrenière A, Weir LK, Bernatchez L. 2012. Framing the salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS One. 7:e46662
- Doiron S, Bernatchez L, Blier PU. 2002. A comparative mitogenomic analysis of the potential adaptive value of Arctic charr mtDNA introgression in brook charr populations (Salvelinus fontinalis Mitchill). Mol Biol Evol. 19:1902–1909.
- Frolov SV, Frolova VN. 2001. The karyotype of Chukotka chars from the Estikhed Lake (Eastern Chukotka). Russ J Genet. 37:180–183.
- Glubokovsky MK, Chereshnev IA, Chernenko EV, Viktorovsky RM. 1979. Distribution of the arctic charr group (Salvelinus, Salmoniformes) along the Asiatic Pacific coast. In: Systematics and ecology of fishes in continental water bodies of the Far East. Vladivostok: Far East Science Center, USSR Academy of Sciences; p. 86–98.
- Oleinik AG, Skurikhina LA, Brykov VA. 2015. Phylogeny of charrs of the genus Salvelinus based on mitochondrial DNA data. Russ J Genet. 51:55–68.
- Omel’chenko VT, Salmenkova EA, Malinina TV, Frolov SV. 1998. Genetic differentiation of sympatric populations of chars of the genus Salvelinus from the Achchen Lake (Chukotka). Russ J Genet. 34:311–316.
- Yamamoto S, Maekawa K, Morita K, Crane P, Oleinik A. 2014. Phylogeography of the salmonid fish, Dolly Varden Salvelinus malma: multiple glacial refugia in the North Pacific Rim. Zool Sci. 31:660–671.
- Uiblein F, Jagsch A, Honsig-Erlenburg W, Weiss S. 2001. Status, habitat use, and vulnerability of the European grayling in Austrian waters. J Fish Biol. 59(Supple. A):223–247.
- Ward RD, Zemlak TS, Innes BH, Last PR, Hébert PD. 2005. DNA barcoding Australia's fish species. Philos Trans R Soc Lond B Biol Sci. 360:1847–1857.
