Abstract
The mitochondrial genome of the hog deer (Axis porcinus) was sequenced using an Illumina MiSeq. The assembled genome consists of 16,351 bp, and shared a 99.8% similarity to the published chital deer (Axis axis) genome, suggesting that they belong to the same species. Further research is ongoing to understand why these mitochondrial genomes are highly similar.
The hog deer (Axis porcinus) was introduced into Australia in the nineteenth century by Acclimatisation Societies, primarily for hunting purposes (Mayze and Moore Citation1990). It is now endangered throughout its native range of Pakistan, India, Nepal, and Bangladesh, however, a stable population exists in the Gippsland region of Victoria, Australia (Scroggie et al. Citation2012). There is currently limited genetic knowledge of the population diversity, structure, and origin of the Australian A. porcinus population.
Presented here is the mitochondrial genome of A. porcinus from Victoria, Australia, sequenced via next generation sequencing methods. Liver samples were collected from four hog deer on Sunday Island, Victoria, Australia during the 2015 hog deer hunting season (−38.711683° 146.623333°). Genomic DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. NebNext Ultra Library Prep Kit (BioLabs, Ipswich, MA) was used to create pair-end libraries, with size selection of 500–600 bp. The MiSeq Reagent Kit v3 (2x300) (Illumina, San Diego, CA) was used to prepare samples for sequencing on an Illumina MiSeq. The reads obtained from the MiSeq run were then processed using the program Kraken v0.10.5 (Wood and Salzberg Citation2014) to remove non-target DNA sequences, resulting in an average of 5 million reads per sample. CLC Genomics Workbench 7 (CLC bio, Inc., Aarhus, Denmark) was used for de novo assembly, with a minimum contig size of 1000. Protein coding regions and ribosomal sequences were determined using MITOS (Bernt et al. Citation2013).
Mitochondrial genomes of 12 closely related deer species were downloaded from GenBank, in addition to the A. porcinus mitochondrial genome from Australia. Phylogenetic analyses were performed using the maximum-likelihood method. All genomes were aligned using Geneious 9.0.5 (Kearse et al. Citation2012). The best-fit model GTR + G was determined using JModelTest 2.1.7 (Darriba et al. Citation2012), under the Akaike Information Criterion (Akaike Citation1974). A phylogenetic tree was created using the program RAxML (Stamatakis Citation2014), with 1000 bootstrap replicates. The Muntiacus muntjak (AY225986) mitochondrial genome was used as an outgroup.
The length of the A. porcinus genome was 16,351 bp with the base composition of A (33.4%), T (29.3%), C (24%), and G (13.3%) (GenBank Accession no. MF435989). The genome includes 13 protein coding genes, two rRNA genes, and 22 tRNA genes.
The phylogenetic tree shows that the Victorian A. porcinus and A. axis (JN632599) form their own clade, separate from the other A. porcinus genome (JN632600) (). Bootstrap values show 100% support for both Axis clades.
Figure 1. Maximum likelihood phylogenetic tree of complete mitochondrial genomes of Victorian A. porcinus and closely related species. GenBank accession numbers are listed next to species’ names. Bootstrap values are listed next to nodes, with 1000 replicates. Muntiacus muntjak was used as an outgroup.
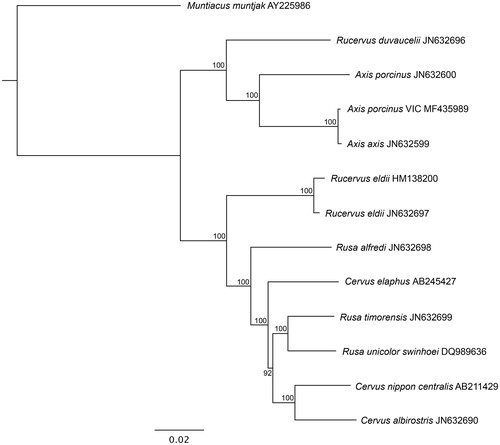
The Victorian A. porcinus shared a 99.8% similarity to the mitochondrial genome of A. axis, and only 94.66% similarity to another A. porcinus genome. In comparison, two mitochondrial genomes of Rucervus eldii taken from GenBank (HM138200 and JN632697) shared a 99.4% similarity.
No samples of chital were analysed in the lab prior to this experiment, so contamination of samples can be ruled out. Further research is ongoing to understand why the mitochondrial genomes of Victorian A. porcinus and A. axis are so similar.
Acknowledgements
This project was jointly funded by the RFA grant ‘Securing Food, Water and the Environment’ (La Trobe University), and the Victorian Game Management Authority. We thank Steve Doyle and Jude Hatley for their assistance in preparing samples to be run on the Illumina MiSeq. We also thank hunters who provided access to samples and to Victorian Hog Deer Checking Station operators and Game Management Authority staff for assisting with the collection of samples.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Autom Control. 19:716–723.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Mayze RJ, Moore G. 1990. The hog deer. Croydon (VIC). Australian Deer Research Foundation.
- Scroggie MP, Forsyth DM, Brumley AR. 2012. Analyses of Victorian hog deer (Axis porcinus) checking station data: demographics, body condition and time of harvest. Arthur Rylah Institute of Environmental Research Technical Report Series. No. 230.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312?1313.
- Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15:R46.
