Abstract
Gymnodraco acuticepsis is an Antarctic fish living in the Southern Ocean. Until now, studies on G. acuticeps are still limited. As an Antarctic fish, obtaining and characterization of the mitochondrial genome of G. acuticeps will be important for elucidation of the mechanism of cold-adapting evolution in mitochondrion. In this study, we first isolated and characterized the mitochondrial genome sequence of G. acuticeps with 15,987 bp in length. It contained of 34 genes (12 protein-coding genes, 20 transfer RNA genes, 2 ribosomal RNA genes) and a partial putative control region. Gene organization and nucleotide composition of obtained mito-genome were similar to those of other Antarctic fish. Twenty-eight genes were encoded by heavy strand, while six genes were encoded by light strand. Further, the phylogenetic tree, which based on 12 protein-coding genes, revealed that the G. acuticeps was genetically closest to species Parachaenichthys charcoti among 18 species. We hope this work would be helpful for the population genetics and molecular evolution studies.
The family Bathydraconidae of the suborder Notothenioidei was consisted of at least 11 genera and 16 species living in the south of the Antarctic Polar Front. Gymnodraco acuticeps belongs to family Bathydraconidae which lived beneath the sea ice of the Southern Ocean (Eastman and Hikida Citation1991). It experience unusual environmental conditions, including highly oxygenated subzero water. The common name of G. acuticeps is the naked dragonfish, which stems from its body feature such as lack of scales, the dragon-like form of its head, a protruding lower jaw bearing prominent, and the exposed canines. They are the predators in Antarctic food web, feeding on a variety of organisms including some fish, amphipods and polychaetes (Evans et al. Citation2005). Some regions of the mitochondrial genome were thought to be the ideal markers for studies on population genetic diversity, molecular phylogeny, and species identification due to the high mutation rate, simple structure, abundant distribution and maternal inheritance. So far, mitochondrial genome sequence of G. acuticeps is still unavailable and this has hampered the genetic study in this group fishes.
Adult fish of G. acuticeps was collected near Zhongshan Station (68tio 708tio after freezing at −80 °C), it was transported to East China Sea Fisheries Research Institue, Chinese Academy of Fishery Science for storage and DNA extraction. In this study, we determined the nearly complete mitochondrial genome of G. acuticeps (the accession number: KX840362). This genome was 15,987 bp in length, including 12 protein-coding genes, 20 tRNA genes, two rRNA genes. A non-coding region with high A + T content between tRNAPro and tRNAPhe was identified as the putative control region. Recently, the mitochondrial gene such as ND6 and tRNAGlu were found to be ‘lost’ in Antarctic notothenioids (Papetti et al. Citation2007). Later they were found between duplicated control regions (CRs) in rearranged mitochondrial genomes of this taxon (Zhuang and Cheng Citation2010). The structure and gene arrangement of obtained mitochondrial genome of G. acuticeps were similar with those obtained from other fish species such as Chionodraco hamatus (Song et al. Citation2016), Notothenia coriiceps and Notothenia rossii (Zhuang and Cheng Citation2010). The total nucleotide composition of G. acutips is mitogenome was 25.11% for A, 17.40% for G, 30.80% for C, and 26.69% for T, with a high A + T content of 51.80%. Twelve protein-coding genes were 10,948 bp in length and the composition was 23.44% for A, 16.72% for G, 31.91% for C and 27.93% for T, with an A + T content of 51.37%.
Among 34 genes, 28 were encoded by the heavy strand (H-strand) and only six genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr and tRNASer) were encoded by light strand (L-strand). 11 protein-coding genes were started by ATG, while the other protein-coding gene COI was initiated by GTG. In total, four types of stop codons (TAG, TAA, TAG, and T-) were detected in these 12 protein-coding genes, and the gene ND5 was stopped by a rare stop codon TAG. A total of 14 intergenic spacers were found in this genome, 10 on H-strand, and four on L-strand with the ranges from 1 to 44 bp. Meanwhile, six overlaps were observed, with the lengths between 1 to 10 bp. The longest overlap (10 bp) occurred between ATP8 and ATP6, while the biggest intergenic spacer (44 bp) was located between tRNAAsn and tRNACys. The putative origin of light strand replication was found between tRNAAsn and tRNACys, with a length of 44 bp.
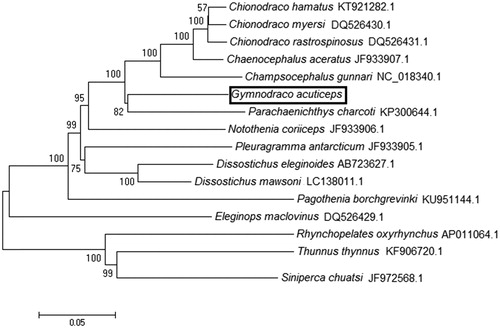
The phylogenetic relationship of G. acuticeps within 13 Antarctic fish was analyzed using 12 concatenated protein-coding genes except ND6 (). Usually, ND6 was not used for phylogenetic analysis due to its high heterogeneity and poor phylogenetic performance (Miya and Nishida Citation2000). The phylogenetic tree was reconstructed using neighbor-joining (NJ) algorithms in MEGA 4.0 software with 1000 bootstrap replicates (Zardoya and Meyer, Citation1996). From the tree topologies, we can find that the G. acuticeps was genetically closest to species Parachaenichthys charcoti among 16 species. This result is identical to previous phylogeny studies by using the partial mitochondrial gene (Bargelloni et al. Citation1994).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bargelloni L, Ritchie PA, Patarnello T, Battaglia B, Lambert DM, Meyer A. 1994. Molecular evolution at subzero temperatures: mitochondrial and nuclear phylogenies of fishes from Antarctica (suborder Notothenioidei), and the evolution of antifreeze glycopeptides. Mol Bio Evol. 11:854–863.
- Eastman JT, Hikida RS. 1991. Skin structure and vascularization in the Antarctic notothenioid fish Gymnodraco acuticeps. J Morphol. 208:347–365.
- Evans CW, Cziko P, Cheng CHC, Devries AL. 2005. Spawning behaviour and early development in the naked dragonfish Gymnodraco acuticeps. Antarct Sci. 17:319–327.
- Miya M, Nishida M. 2000. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 17:437.
- Papetti C, Liò P, Rüber L, Patarnello T, Zardoya R. 2007. Antarctic Fish Mitochondrial Genomes Lack ND6 gene. J Mol Evol. 65:519–528.
- Song W, Li LZ, Huang HL, Meng YY, Jiang KJ, Zhang FY, Chen XZ, Ma LB. 2016. The complete mitochondrial genome of Chionodraco hamatus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B: Resources. 1:52–53.
- Zardoya R, Meyer A. 1996. The complete nucleotide sequence of the mitchondrial genome of the lungfish (Protopterus dolloi) supports its phylogenetic position as a close relative of land vertibrates. Genet. 142:1249–1263.
- Zhuang X, Cheng CH. 2010. ND6 gene ‘lost and found: evolution of mitochondrial gene rearrangement in Antarctic notothenioids’. Mol Biol Evol. 27:1391.