Abstract
The complete mitochondrial genome of Electrona carlsbergi was obtained, which was 18,282 bp in size and including 13 protein-coding genes, 2 ribosomal RNAs, 23 transfer RNAs and 1 control region. The overall nucleotide composition is 27.92% for A, 24.66% for T, 30.90% for C and 16.52% for G. Among 23 tRNA genes, 8 tRNAs were encoded on the L-strand. Further, the phylogenetic tree, which based on complete mtDNA sequences, revealed that the E. carlsbergi was genetically closest to species E. antarctica and Krefftichthys anderssoni. This study could provide a basic data for the studies on evolution for low temperature adaptability, stock evaluation and conservation genetics.
In terms of biomass level, Electrona is the second largest marine organism in Antarctic Ocean and including three species of Electrona, like Electrona antarctica, Electrona carlsbergi and Electrona rissoi (Zhu et al. Citation2012). E. carlsbergi (Myctophidae, Electrona), appeared in the late Miocene, is one of the most abundant species in Antarctic waters. They cover waters from the south of the Antarctic convergence to the Antarctic coast and generally live in the depth of 200–400 m of water. The optimum water temperature is 1°C (Cheung et al. Citation2013). As an important member of the Antarctic ecosystem, E. carlsbergi mainly preys on Antarctic krill, copepods, and cephalopods. The distribution of E. carlsbergi is most affected by temperature. Their maximum length is 10.5 cm and the body of females are larger than males. So far, no complete mitochondrial sequence information is available. The study is important for the Antarctic ecosystem and further research on genetics and evolution of E. carlsbergi.
Adult fish of E. carlsbergi was collected from Antarctic (63°14′54″S, 59°51′24″W), it was transported to East China Sea Fisheries Research Institute, Chinese Academy of Fishery Science after freezing at −80°C. The genomic DNA was extracted from muscle tissues using Animal Genomic DNA Extraction Kit (TIANGEN) following the operation manual. The amplifying and sequencing primers were designed according to the sequence of E. antarctica (AP012248.1). We acquired the complete mitochondrial sequence of E. carlsbergi and submitted it into the Genbank database with an accession number MF596172. This complete mitochondrial genome is 18,282 bp in length, including 13 protein-coding genes, 2 rRNA genes, 23 tRNA genes and 1 control region. The overall nucleotide composition is 27.92% for A, 24.66% for T, 30.90% for C and 16.52% for G. The GC content (47.42%) is similar to Chionodraco hamatus (Song et al. Citation2016). In 13 protein-coding genes, there types of initiation codon (ATC, ATG, GTG) were identified. Four types of stop codons (TAA, TAG, TA, T) were detected and the rare stop codon TAG stopped ND3, it was different from Gymnodraco acuticeps. Eight of 23 tRNAs were encoded on the L-strand. The length of control region (D-loop) is 1990bp, longer than other fishes and its overall nucleotide composition is 35.23% for A, 31.81% for T, 21.61% for C and 11.36% for G. It has many long tandem repeat, like (ATTATACCCATAACTTGATATAACCC)8 and (ATATGTATTATACCC)12.
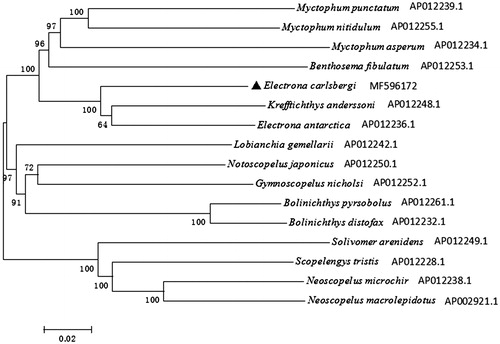
To evaluate the phylogenetic position of E. carlsbergi in Myctophidae fishes, the phylogenetic tree was reconstructed based on complete mtDNA sequences using the Neighbour-joining method in MEGA 5.1 (Kumar et al. Citation2008) (). Neoscopelus microchir, Neoscopelus macrolepidotus, Scopelengys tristis and Solivomer arenidens were used as an out-group. The NJ tree showed that E. carlsbergi clustered with E. antarctica and Krefftichthys anderssoni, then together with other species in family Myctophidae, forming a big branch. Besides, the out-group in family Neoscopelidae formed a big sister branch as well.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. This work was supported by the Sub-project under National Science & Technology Support Plan (No. 2013BAD13B03), and the Chinese National Antarctic Research Expedition (CHINARE2014-01-06).
References
- Cheung WWL, Watson R, Pauly D. 2013. Signature of ocean warming in global fisheries catch. Nature. 497:365.
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatics. 9:299–306.
- Song W, Li LZ, Huang HL, Meng YY, Jiang KJ, Zhang FY, Chen XZ, Ma LB. 2016. The complete mitochondrial genome of Chionodraco hamatus (Notothenioidei: Channichthyidae) with phylogenetic consideration. Mitochondrial DNA Part B:Resources. 1:52–53.
- Zhu G, Li F, Feng C, Wu Q, Xu P, Xu L. 2012. The biology of electrona antarctica in the western south orkney islands, antarctic. Chinese J Polar Res. 24:346–351.