Abstract
Antheraea frithi mitogenome was sequenced to understand its phylogenetic and evolutionary relationship. It comprises 22 tRNAs, 13 protein-coding genes, two rRNAs and a AT rich region. The arrangement of mitogenome gene follows the pattern of dytrysian lepidoptera. rrnL gene consists of two unique long consensus repeats copy. tRNAs like structure of trnL and trnP are present in rrnL gene and AT rich region, respectively. The phylogenetic analysis by maximum-likelihood method showed that A. frithi was clustered with A. pernyi and A. yamamai, the commercialized wild silkworms.
Antheraea frithi, the oak tasar silkworm, is an unexplored wild sericigenous insect that showed commercial potentiality in Manipur. Proper biological and genetic characterizations of A. frithi are essential for its successful conservation and commercialization. The complete mitochondrial genome of A. frithi was determined to examine its comprehensive evolutionary relationship and maternal inheritance among sericigenous groups.
Live samples of A. frithi were collected from Phaibung (1200 m ASL), Senapati district (N25°42′, E94°12′), Manipur, India. Taxonomically identified A. frithi was stored in the IBP, IBSD under the number WS/IBSD/008. The silk gland was used for extraction of mitochondrial DNA (mtDNA). mtDNA was extracted following the protocol of Clark and Nicklas (Citation1970) with slight modification. The isolated mtDNA was sequenced using Illumina HiSeq 2000. PCGs of the mitogenome were identified by ORF finder. tRNA genes were identified in tRNAscan-SE Search (http://lowelab.ucsc.edu/tRNAscan-SE/). Phylogenetic trees based on maximum likelihood analysis were constructed on MEGA 5.1 (Tamura et al. Citation2011).
Mitogenome of A. frithi (GenBank KJ740437) was 15,338 bp in length. It contains typical composition of 37 genes found almost in mitochondrial genomes of insects, having 13 protein coding genes (PCGs), 22 tRNA genes, two subunit of rrnS and rrnL, and AT-rich region. The mitogenome follows orientation and gene order of ditrysian species of Lepidoptera (trnM, trnI, trnQ) (Chen et al. Citation2014). Within 13 PCGs, AT composition was maximum in the atp8 gene (91.51%) and minimum in the cox1 gene (72.15%). Antheraea frithi has high AT% (89.19%) in the control region as similar with Artogeia melete (89.17%) (Hong et al. Citation2009). rrnL consists of two long consensuses repeats of 42 bp and 32 bp. A unique feature of trnL like structure is its presence in rrnL gene. In the AT rich region, there is a structure with the motif ‘ATAGA’ and 19 bp poly (T) stretches at the 18 bp downstream of rrnS gene. trnP-like structure having proper anticodon but partial DHU loop is also detected.
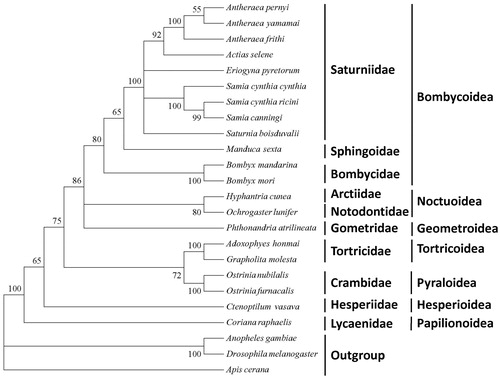
The phylogenetic tree of A. frithi, built from the 13 PCGs of mitogenomes shows clustering of A. pernyi and A. yamamai in one distinct clade. Within Bombycoidea superfamily, Bombycidae, Saturniidae and Sphingoidae designated a cluster following the traditional morphology and molecular based classification (Liu et al. Citation2013). The superfamily, Papilionoidea forms a sister clade to Lepidopteran superfamilies (Liu et al. Citation2012). The study provides fundamental data useful in conservation genetics and thriving the commercial aspect among the wild silkworm diversification.
Figure 1. Maximum-likelihood tree of A. frithi with other related species based on 13 protein-coding genes. The sequences accession number of the species used in phylogenetic analysis are listed as follows: Antheraea pernyi (HQ264055); Antheraea yamamai (EU726630); Actias selene (NC_018133); Eriogyna pyretorum (NC_012727); Samia cynthia cynthia (NC_002084); Samai cynthia ricini (AJ400907); Samia canningi (NC_016704); Saturnia boisduvalli (NC_010613.1), Manduca sexta (NC_003368); Bombyx mandarina (NC_024270); Bombyx mori (NC_010266); Hyphantria cunea (AM946601); Ochrogaster lunifer (NC_003367); Phthonandria atrilineata (NC_007976); Adoxophyes honmai (NC_010613); Grapholita molesta (NC_014058); Ostrinia nubilalis (NC_008141); Ostrinia furnacalis (HQ116416); Ctenoptilum vasava (AB070264); Coreana raphaelis (NC_003395); Anopheles gambiae (KC812618); Drosophila melanogaster (NC_010522); Apis cerana (NC_017869).
Acknowledgements
The authors also thank the Director, IBSD, Manipur, India for providing laboratory facilities for the execution of this study.
Disclosure statement
The authors report that they have no competing interest.
Additional information
Funding
References
- Chen MM, Li Y, Chen M, Wang H, Li Q, Xia R, Zeng CY, Li YP, Liu YQ, Qin L. 2014. Complete mitochondrial genome of the atlas moth, Attacus atlas (Lepidoptera: Saturniidae) and the phylogenetic relationship of Saturniidae species. Gene. 545:95–101.
- Clark JB, Nicklas WJ. 1970. The metabolism of rat brain mitochondria. Preparation and characterization. J Biol Chem. 245:4724–4731.
- Hong G, Jiang S, Yu M, Yang Y, Li F, Xue F, Wei Z. 2009. The complete nucleotide sequence of the mitochondrial genome of the cabbage butterfly, Artogeia melete (Lepidoptera: Pieridae). Acta Biochim Biophys Sin (Shanghai). 41:446–455.
- Liu QN, Zhu BJ, Dai LS, Liu CL. 2013. The complete mitogenome of Bombyx mori strain Dazao (Lepidoptera: Bombycidae) and comparison with other lepidopteran insects. Genomics. 101:64–73.
- Liu QN, Zhu BJ, Dai LS, Wei GQ, Liu CL. 2012. The complete mitochondrial genome of the wild silkworm moth, Actias selene. Gene. 505:291–299.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
