Abstract
The complete mitochondrial genome of marine fish Pelates quadrilineatus was sequenced by the high throughput sequencing method. This genome was 16,823 bp in length, consisted of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one large non-coding region. The gene arrangement of P. quadrilineatus is identical to those of other fishes. Phylogenetic tree based on 13 protein coding genes shows that Terapontidae has a closer phylogenetic relationship to Pentacerotidae than to Chaetodontidae.
Pelates quadrilineatus, common name Fourlined Terapon, inhabits in brackish waters (Kuiter and Tonozuka Citation2001) and feeds on small fishes and invertebrates. Eggs of this species are hatched by the male parent (Breder and Rosen Citation1966), juveniles usually live in seagrass beds or mangrove bays (Kuiter and Tonozuka Citation2001). Now P. quadrilineatus has been classified into family Terapontidae, order Perciformes (Nelson et al. Citation2016), because the morphological differences between Terapontidae fishes are not obvious. The fringes of body side, which is main identified character of P. quadrilineatus, change greatly at different stages of maturity (Vari Citation1978). This phenomenon has made it necessary to develop molecular markers to figure out controversial issues about phylogenetic relationship of P. quadrilineatus (Paxton Citation1989; Faith et al. Citation2004). In this study, we first reported the complete mitochondrial genome of P. quadrilineatus, and analyzed its phylogenetic relationship with some other species from family Pentacerotidae and Chaetodontidae, based on samples collected from Naozhou island in Zhanjiang, China (geographic coordinate: N 20°53′20.11″, E 112°28′46.2″). The whole body specimen was preserved in ethanol and registered to the Marine Biodiversity Collection of South China Sea, Chinese Academy of Sciences, under the voucher number SCF20171022001.
The complete mitochondrial genome of P. quadrilineatus was 16,823 bp in length (GenBank accession no. MG271911), including 13 protein-coding genes, two rRNA genes, 22 tRNA genes, one OL (origin of replication on the light-strand) and one D-Loop (control region). The OL was 51 bp in length, located in the cluster of five tRNA genes (WANCY region) between tRNA-Asn and tRNA-Cys. The D-loop was 956 bp in length, located between tRNA-Pro and tRNA-Phe. Gene arrangement of this genome was identical to those of other fish, and most genes in this genome were encoded by the heavy strand (H-strand), except for ND6 and eight tRNA genes (Boore Citation1999; Yu and Kwak Citation2015; Gong et al. Citation2017). Overall base composition values for the mitochondrial genome were 27.3%, 30.2%, 16.7%, and 25.8% for A, C, G, and T, respectively.
The phylogenetic relationships of P. quadrilineatus with 11 closely related species were analyzed in this study. Complete mitochondrial genes of these 11 species were available on GenBank. The maximum-likelihood evolutionary tree (ML tree) was constructed by MEGA 7 (Kumar et al. Citation2016) based on 1st and 2nd codon sequences of 13 protein coding genes.
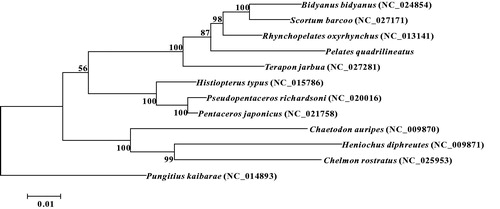
In the ML phylogenetic tree, P. quadrilineatus clustered with Terapon jarbua, Rhynchopelates oxyrhynchus, Scortum barcoo and Bidyanus bidyanus with a strong support. And these five species were all classified into family Terapontidae of order Perciformes. As a sister lineage to the former mentioned clade, Histiopterus typus, Pseudopentaceros richardsoni and Pentaceros japonicus formed another clade, which was classified into family Pentacerotidae of order Perciformes. Another clade included Chaetodon auripes, Heniochus diphreutes and Chelmon rostratus with a strong support, which was classified into family Chaetodontidae of order Perciformes (). These results show that Terapontidae has a closer phylogenetic relationship to Pentacerotidae than to Chaetodontidae.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Breder CM, Rosen DE. 1966. Modes of reproduction in fishes. Q Rev Biol. 155:302–301.
- Faith DP, Reid CAM, Hunter J. 2004. Integrating phylogenetic diversity, complementarity, and endemism for conservation assessment. Conserv Biol. 18:255–261.
- Gong L, Liu LQ, Guo BY, Ye YY, Lü ZM. 2017. The complete mitochondrial genome characterization of Thunnus obesus (Scombriformes: Scombridae) and phylogenetic analyses of Thunnus. Conserv Genet Resour. 9:1–5.
- Kuiter RH, Tonozuka T. 2001. Pictorial guide to Indonesian reef fishes. Part 1 eels-snappers, Muraenidae-Lutjanidae.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Nelson JS, Grande T, Wilson MVH. 2016. Fishes of the World, 5th edition. Hoboken (NJ): John Wiley & Sons, Inc.
- Paxton JR. 1989. Pisces: Petromyzontidae to carangidae. Australian Government Pub.
- Vari RP. 1978. The terapon perches (Percoidei, Teraponidae): a cladistic analysis and taxonomic revision. Bulletin of the AMNH. v. 159, article 5.
- Yu JN, Kwak M. 2015. The complete mitochondrial genome of Brachymystax lenok tsinlingensis (Salmoninae, Salmonidae) and its intraspecific variation. Gene. 573:246–253.