Abstract
The mitochondrial genome of Atrocalopteryx melli was sequenced and assembled via Next-Generation Sequencing (NGS) and iteratively assembly process with a reference seed. This genome is 15,562 bp long and A + T biased (71%), with 37 genes arranged in common order of Odonata. All protein-coding genes are initiated by typical “ATN” codon, and 9 genes are terminated with a complete stop codon, except nad4, nad5, cox2, and cox3, which are terminated with an incomplete codon “T(aa)”. The S5 intergenic spacer is absent in this genome, supporting that lacking of S5 as a specific character for damselflies. The A + T rich region of A. melli is 267 bp longer than that of A. atrata. This mitogenome provides new molecular information for understanding of A. melli and Atrocalopteryx.
Atrocalopteryx, once classified into Calopteryx, has already been separated from the genus Calopteryx by using molecular phylogenetic method (Dumont et al. Citation2005). Even in Atrocalopteryx, a recent extended phylogeny revealed that one species, A. oberthueri, is different from the other species, and it was suggested to be picked out as a new genus (Dumont et al. Citation2007; Guan et al. Citation2012). Thus, Atrocalopteryx could be split into two genera at least, but its true phylogeny has not been well-resolved yet. More detailed molecular information such as mitochondrial genome and data from more species is required to figure out the reliable phylogeny and infer evolutionary history of Atrocaloptryx (Graybeal Citation1998; Philippe et al. Citation2011; Schreeg et al. Citation2016). Atrocalopteryx melli (Ris Citation1912), a large Chinese endemic and beautiful species, is locally abundant in Southern China: Guandong, Fujian, and Guangxi provinces. This species can be found in small shaded headwater streamlets of forests. Here, we sequenced and annotated the mitogenome of A. melli to provide molecular information for genetically understanding of the beautiful animal.
A single individual of A. melli was sampled from a national natural reserve (a member of IBP, 1979) in Dinghushan mountain, Zhao Qing, Guangdong province (N 23°10′21″ E 112°31′39″), and was preserved in Institute of Hydrobiology, Jinan University, Guangzhou, China. The individual was used for genomic DNA extraction with standard phenol-chloroform method described by Hadrys et al. (Citation1992). Genomic DNA was sequenced on an Ion Torrent PGM sequencer in the Research Group Aging Physiology and Molecular Evolution, Ghent University, Belgium. Skewer v.0.2.1 was used to trim all nucleotide with Phred quality scores under 20 (Jiang et al. Citation2014). COI sequence of A. atrata was used as reference seed for iterative assembly by MITObim v.1.8 (Hahn et al. Citation2013). Two primers were designed at both ends of the longest assembled sequence, the PCR amplicon was assembled to the original longest sequence using SeqMan v.7.1.0 (Swindell and Plasterer Citation1997), to get overhang at both ends, which cyclized the final genome with higher reliability. The mitochondrial genome was annotated with the MITOS WebServer (Bernt et al. Citation2013) and verified via BLAST (Altschul et al. Citation1990). Transfer RNA genes were then double-checked with tRNAscan-SE v.2 (Lowe and Eddy Citation1997) and ARWEN v.1.2.3 (Laslett and Canback Citation2008). Finally, an ultrametric Bayesian phylogenetic tree was reconstructed on 13 PCGs of all available mitogenome of Odonata from NCBI and A. melli, following the strategy by Hoi-Sen Yong et al. (Citation2016).
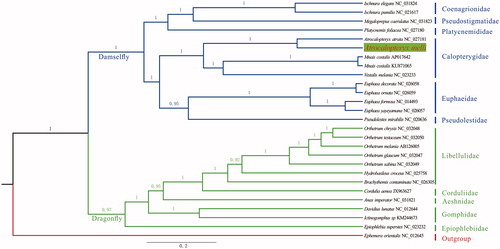
The mitogenome of A. melli is 15,562 bp (GenBank accession number: MG011692) in length, with A + T biased base composition: A (41%), T (30%), G (13%), C (16%). Totally, 37 genes are annotated, comprising 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes, ordered in the common arrangement as other Odonata. All PCGs have “ATN” start codons. Nine PCGs are stopped by complete codon: nad3 and cytb with TAG, nad1, nad2, nad4l, nad6, cox1, atp8, and atp6 with TAA. However, nad4, nad5, cox2, and cox3 end with incomplete codon, posttranscriptional polyadenylation could be indispensable for successful translation. tRNA genes have lengths from 64 bp to 72 bp, and they can be folded in the typical cloverleaf structure. The control region (A + T rich: 79%) has a length of 928 bp, shorter than most other Odonata (Lee et al. Citation2009; Feindt, Herzog, et al. Citation2016; Feindt, Osigus, et al. Citation2016; Herzog et al. Citation2016). Moreover, there are three intergenic spacers with sequence lengths from 9 bp to 16 bp. The S5 spacer is absent in A. melli, supporting the earlier finding of a lack of S5 spacer in damselflies (Lin et al. Citation2010), although this finding could not be universal for that group (Herzog et al. Citation2016). Atrocalopteryx melli and A. atrata (NC_027181.1) have the same mitochondrial genes order and composition. However, A. atrata has a shorter A + T rich region with length of 661 bp. Our reconstructed phylogenetic tree () showed a sister relationship among Atrocalopteryx and Mnais + Vestalis, which is different from Vestalis + (Atrocalopteryx + Mnais) in the phylogeny of Guan et al. (Citation2012). In contrast to mitochondrial PCGs in the present analysis, nuclear sequences (ITS and 18S rDNA) were used in Guan et al.’s phylogeny, which could be the cause for the above-mentioned difference. Hence, except from getting whole mitochondrial genome and data from more species, combining molecular information from mitochondrion and nuclear might be necessary to fully and reliably resolve phylogenetic relationship within Calopterygidae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bernt M, Donath A, Juehling F, Externbrink F, Florentz C, Fritzsch G, Puetz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dumont HJ, Vanfleteren JR, De Jonckheere JF, Ph HW. 2005. Phylogenetic relationships, divergence time estimation, and global biogeographic patterns of calopterygoid damselflies (Odonata, Zygoptera) inferred from ribosomal DNA sequences. Syst Biol. 54:347–362.
- Dumont HJ, Vierstraete A, Vanfleteren JR. 2007. A revised molecular phylogeny of the Calopteryginae (Zygoptera: Calopterygidae). Odonatologica. 36:365–372.
- Feindt W, Herzog R, Osigus HJ, Schierwater B, Hadrys H. 2016. Short read sequencing assembly revealed the complete mitochondrial genome of Ischnura elegans Vander Linden, 1820 (Odonata: Zygoptera). Mitochondrial DNA B Resour. 1:574–576.
- Feindt W, Osigus HJ, Herzog R, Mason CE, Hadrys H. 2016. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA B Resour. 1:497–499.
- Graybeal A. 1998. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 47:9–17.
- Guan Z, Han BP, Vierstraete A, Dumont HJ. 2012. Additions and refinements to the molecular phylogeny of the Calopteryginae S.L. (Zygoptera: Calopterygidae). Odonatologica. 41:17–24.
- Hadrys H, Balick M, Schierwater B. 1992. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1:55–63.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator Leach, 1815 (Odonata: Aeshnidae) via NGS sequencing. Mitochondrial DNA B Resour. 1:783–786.
- Jiang H, Lei R, Ding S-W, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 15:182.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lee EM, Hong MY, Kim MI, Kim MJ, Park HC, Kim KY, Lee IH, Bae CH, Jin BR, Kim I. 2009. The complete mitogenome sequences of the palaeopteran insects Ephemera orientalis (Ephemeroptera: Ephemeridae) and Davidius lunatus (Odonata: Gomphidae). Genome. 52:810–817.
- Lin CP, Chen MY, Huang JP. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa support a basal Odonata within the Pterygota. Gene. 468:20–29.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D. 2011. Resolving difficult phylogenetic questions: Why more sequences are not enough? PLoS Biol. 9:e1000602.
- Ris F. 1912. Neue Libellen von Formosa, Sudchina, Tonkin und den Philippinen. Supplementa Entomologica. 1:44–85.
- Schreeg ME, Marr HS, Tarigo JL, Cohn LA, Bird DM, Scholl EH, Levy MG, Wiegmann BM, Birkenheuer AJ. 2016. Mitochondrial genome sequences and structures aid in the resolution of Piroplasmida phylogeny. PLos One. 11:e0165702
- Swindell SR, Plasterer TN. 1997. Sequence Data Analysis Guidebook. In: Swindell SR, editor. Totowa (NJ): Springer; p. 75–89.
- Yong HS, Song SL, Suana IW, Eamsobhana P, Lim PE. 2016. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem Syst Ecol. 69:124–131.