Abstract
The complete mitochondrial genome of Ancherythroculter wangi was determined. The mitochondrial genome, consisting of 16,622 base pairs (bp), encoded 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs and a non-coding control region, as those found in other Ancherythroculter species. These results can provide useful data for further studies on taxonomic status, molecular systematics, and stock evaluation.
Ancherythroculter wangi belongs to the family Cyprinidae and the order Cypriniformes, and mainly distributed in the upper stream of Yangtze River and its tributary (Ding Citation1994; Yue et al. Citation2000). There was less report about its basic biology data including genetic information. In this study, we first determined the complete mitochondrial genome of A. wangi, which provides the basic molecular data for the further study on its systematics and conservation biology.
In the present study, one specimen A. wangi chosen for mitochondrial genome analysis were collected from the downstream of Qi River (N:30°39.534’, E:105°11.893’) (specimen is stored in Aquaculture Department of Sichuan Agricultural University, number Aw2017092801). Primers were designed for polymerase chain reaction (PCR) amplification and sequencing on the basis of the mitogenome sequence of Ancherythroculter lini (GenBank accession no. NC_027741) (Chen et al. Citation2016). The complete mt genome of A. wangi was 16,621 bp and has been deposited in GenBank with accession no. MG575902. The mitochondrial genome encoded 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs and a non-coding control region, as those found in other Ancherythroculter species (Wan et al. Citation2013; Chen et al. Citation2016; Wang et al. Citation2016). The nucleotide composition of the genome of A. wangi is A 31.2%, T 24.9%, G 16.2% and C 27.7%. Except for the nad6 and eight tRNA genes (tRNA-Gln, tRNA-Aln, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-SerUCN, tRNA-Glu and tRNA-Pro) encoded on the light-strand, all others genes were encoded on the heavy-strand. This is a typical gene arrangement conforming to the other Ancherythroculter species and vertebrate consensus (Wan et al. Citation2013; Chen et al. Citation2016; Wang et al. Citation2016).
All genes use ATG as start codon, except cox1 use GTG, which also discovered in other Ancherythroculter species (Wan et al. Citation2013; Chen et al. Citation2016; Wang et al. Citation2016). Most open reading frames ended with two types of complete stop codons TAA and TAG, whereas the other seven genes, ND2, COX2, ATP8, COX3, ND3, ND4 and cytochrome b (CYTB), had an incomplete termination codon T or TA.
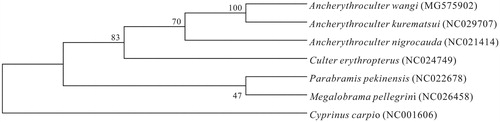
Based on combined nucleotide sequence data of 12 heavy-strand protein-coding genes of A. Wangi, and together with the sequences of other Ancherythroculter species, phylogenetic trees were constructed by using the UPGMA methods (). All Ancherythroculter species had close relationship, A. Wangi, A. kurematsui and A. nigrocaudai were monophyletic in the trees. Numbers on the nodes correspond to bootstrap values based on 1000 iterations. Thus, the mitochondrial genome data and phylogenetic analysis of the A. Wangi can enrich the basic molecular data of Ancherythroculter.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Chen Y, Hu J, Zhao H, Yang H, Liu L. 2016. The complete mitochondrial genome of the Ancherythroculter lini (Cypriniformes, Cyprinidea). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3043–3044.
- Ding RH. 1994. The fishes of Sichuan, China. Chengdu (China): Sichuan Publishing House of Science and Technology. pp. 203–205.
- Yue P, Shan X, Lin R. 2000. Fauna sinica: osteichthyes cypriniformes III. Beijing (China): Science Press.
- Wan Q, Chen Y, Cheng Q, Qiao H. 2013. The complete mitochondrial genome sequence of Ancherythroculter nigrocauda (Cypriniformes:Cyprinidae). Mitochondrial DNA. 24:627–629.
- Wang GJ, Zheng ZL, Yu EM, Xie J, Wei N, Wu JR, Li JS. 2016. The complete mitochondrial genome of Ancherythroculter kurematsui (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 1:630–631.