Abstract
Erythropalum scandens (Erythropalaceae, Santalales) is a large liana distributed in alluvial and riparian forests of Southeast Asia. Here we report and characterize the complete plastid genome sequence of E. scandens in an effort to provide genomic resources useful for developing its medicinal and edible value. The complete plastome is 156,154 bp in length and contains the typical structure and gene content of angiosperm plastomes, including two Inverted Repeat (IR) regions of 26,394 bp, a large single-copy (LSC) region of 84,799 bp and a small single-copy (SSC) region of 18,567 bp. The plastome contains 112 genes, consisting of 79 unique protein-coding genes, 29 unique tRNA genes and four unique rRNA genes. The overall A/T content in the plastome of E. scandens is 62.01%. Phylogenetic analyses were performed using the entire plastome, including genes, spacers and introns, which recovered E. scandens as sister to remaining Santalales with complete plastome sequences.
Erythropalum scandens Blume (Erythropalaceae sensu Nickrent et al. Citation2010) is a large liana endemic to the forests of Hainan Province, China (Qiu and Gilbert Citation2003). It is used as an edible wild vegetable in tropical and subtropical areas of southeast Asia, e.g. local peoples from Guangxi, Guangdong, Hainan and southern Yunnan provinces in China have traditionally collected and eaten its young leaves and stems. It contains resins, phenolic and triterpene medicinal ingredients in the roots. The main components of the resin include sumaresinolic acid and coniferyl cinnamate (Qiu and Gilbert Citation2003). Consequently, genetic and genomic information is urgently needed to promote the medicinal and edible value of E. scandens. Here, we report and characterize the complete plastome of E. scandens (GenBank accession number: this study) based on Illumina paired-end sequencing data (Illumina, San Diego, CA). This is the first report of a complete plastome for the genus Erythropalum and Erythropalaceae, which occupies an important phylogenetic position near the base of the large angiosperm order Santalales (Nickrent et al. Citation2010).
Leaf material of E. scandens was sampled from Diaoluo Mountain National Nature Reserve in Hainan province of China (109.88°E, 18.67°N). A voucher specimen (Wang et al. B40) was deposited in the herbarium of the Institute of Tropical Agriculture and Forestry, Hainan University, Haikou, China. The modified CTAB method of Doyle and Doyle (Citation1987) was used to extract total genomic DNA from leaves quickly frozen with dry ice. 1 μg of genomic DNA was used for Illumina library preparation, using version 3 chemistry. Paired-end, 150 bp reads were sequenced using an Illumina HiSeq 2500 platform at the Guangzhou Novel-seq Biotechnology Co, Ltd (Guangzhou, China). Reads were trimmed and those with >10% Ns or with >10% low-quality (Q ≤ 5) bases were filtered out using NGSQC-Toolkit v2.3.3 (Patel and Jain Citation2012). Cleaned reads were assembled against the plastome of Stephania japonica (NC_029432_1) (Nock et al. Citation2014) using MITO bim v1.8 (Hahn et al. Citation2013).
The plastome was annotated using Geneious R8.0.2 (Biomatters Ltd., Auckland, New Zealand) against the plastome of Stephania japonica. The annotation was corrected with DOGMA (Wyman et al. Citation2004). A circular plastome map was generated using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The plastome of E. scandens was found to possess a total length 156,154 bp with the typical quadripartite structure of angiosperms, containing two Inverted Repeats (IRs) of 26,394 bp separated by a large single-copy (LSC) region and a small single-copy (SSC) region of 84,799 and 18,567 bp, respectively. The plastome was found to contain 125 genes, including 79 protein-coding genes (six of which are duplicated in the IR), four ribosomal RNA genes, and 29 tRNA genes (seven of which are duplicated in the IR). Among these genes, 14 genes (trnA-UGC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, atpF, ndhA, ndhB, petB, petD, rpoC1, rpl2, rpl16, rps16) possessed a single intron and three genes (ycf3, clpP, rps12) had two introns. The gene rps12 was found to be trans-spliced, as is typical of angiosperms. The overall A/T content of the plastome was 62.01%, while the corresponding values of the LSC, SSC and IR regions were 63.77%, 67.66% and 57.18%, respectively.
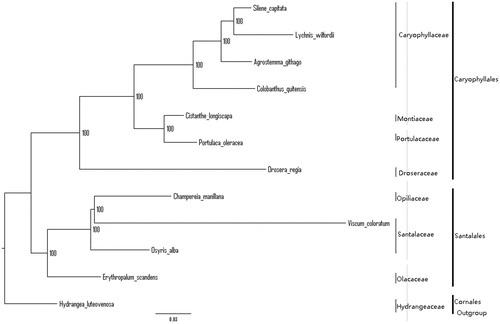
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum-likelihood (ML) phylogeny of 11 published complete plastomes of Santalales and Caryophyllales, using Hydrangea luteovenosa (Hydrangeaceae, Cornales) as an outgroup. The phylogenetic analysis recovered E. scandens as sister to remaining Santalales, with maximum bootstrap support (). The plastome reported here will provide a useful resource for development of the medicinal and edible value of E. scandens, as well as for phylogenetic studies of Santalales.
Figure 1. The best ML phylogeny recovered from plastomes included in this study. Accession numbers: Erythropalum scandens (This study), Osyris alba NC_027960.1, Viscum coloratum NC_035414.1, Champereia manillana NC_034931.1, Agrostemma githago NC_023357.1, Colobanthus quitensis NC_028080.1, Lychnis wilfordii NC_035225.1, Silene capitata NC_035226.1, Drosera regia NC_035415.1, Cistanthe longiscapa NC_035140.1, Portulaca oleracea NC_036236.1, Hydrangea luteovenosa NC_035662.1 (lower in the figure).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome-DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Nickrent DL, Malécot V, Vidal-Russell R, Der JP. 2010. A revised classification of Santalales. Taxon. 59:538–558.
- Nock CJ, Baten A, King GJ. 2014. Complete chloroplast genome of Macadamia integrifolia confirms the position of the Gondwanan early-diverging eudicot family Proteaceae. BMC Genomics. 15 (Suppl. 9):S13.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619
- Qiu HX, Gilbert MG. (2003). Flora of China. Olacaceae. Missouri: Science Press and the Missouri Botanical Garden Press.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
