Abstract
Here, we report the complete mitochondrial genome of Bombyx mandarina strain Shenyang. The genome is 15,682 bp in length and contains a typically conserved 37 metazoan genes (13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes and an A + T-rich region). The A + T-rich region of B. mandarina Shenyang contains only one copy of the repeat element of a 126-bp as found in Chinese B. mandarina. A phylogenetic analysis provides a solid evidence that B. mandarina Shenyang belongs to Chinese B. mandarina, rather than Japanese B. mamdarina. The mitochondrial genome will provide useful information for the origin of the present day silkworm.
The wild silkworm Bombyx mandarina is the wild ancestor nearest to the domestic silkworm Bombyx mori. The wild silkworm is widespread throughout areas of Asia and exhibits great diversity. There exists two types of wild silkworms, Chinese B. mandarina and Japanese B. mamdarina (Astaurov and Rovinskaya Citation1959; Shimada et al. Citation1995). To date, the complete mitogenomes (mitogenome) of 13 Chinese wild silkworms have been available (Pan et al. Citation2008; Hu et al. Citation2010; Li et al. Citation2010); however, only one is from northern China. The representatives distributed in northern China remain severely limited. In the present study, we report the complete mitogenome sequence of B. mandarina strain Shenyang, thus providing a solid evidence that B. mandarina Shenyang belongs to Chinese B. mandarina.
We got larvae of B. mandarina Shenyang at the mulberry field of Shenyang Agricultural University (N41°49′0.32′′; E123°33′3.00′′) in June, 2016. A single larva was used to extract total genomic DNA that was stored in our lab. The whole mitogenome of B. mandarina Shenyang was amplified in 19 overlapping fragments. Protein-coding genes (PCGs), ribosomal RNA genes (rRNAs) and transfer RNA genes (tRNAs) were identified with known B. mandarina mitogenome sequences.
Whole mitogenome sequence of B. mandarina Shenyang is 15,682 bp in length, and has been deposited in GenBank under accession no. MG604734. This mitogenome shares a typical metazoan genes, including 13 PCGs, 22 tRNA genes, two rRNAs, and a non-coding A + T-rich region, exhibiting a typical gene complement, order, and arrangement identical to that of in B. mandarina previously sequenced. Among 13 PCGs, 12 have a typical ATN start codon, and the remaining one COI begins with atypical codon CGA as found in B. mandarina Qingzhou (Hu et al. Citation2010). Ten of 13 PCGs harbor a complete termination codon TAA, but the remaining three possess the incomplete termination codons (two T and one TA). The 22 tRNAs range from 64 bp of tRNAArg to 75 bp of tRNAVal in size. A repeat element of a 126-bp has been identified in Japanese B. mandarina where the 747-bp A + T-rich region contains a tandem triplication (Yukuhiro et al. Citation2002). As found in the A + T rich region of Chinese B. mandarina (Pan et al. Citation2008), only one copy of this repeat element is present in the A + T-rich region of B. mandarina Shenyang.
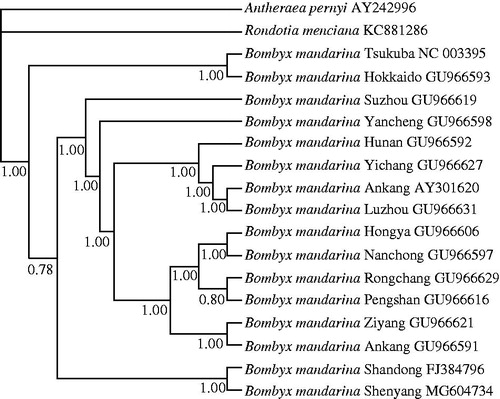
Mrbayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) was used to construct a phylogenetic tree based on whole genome sequences including 16 available strains of B. mandarina. The mitogenome sequences of Rondotia menciana (Kong and Yang Citation2015) and Antheraea pernyi (Liu et al. Citation2008) served as outgroups. Bootstrap values were high for all nodes in the phylogenetic tree. Bayesian inference analysis provided a solid evidence that B. mandarina Shenyang belongs to Chinese B. mandarina (). Geographically, Shenyang is close to southern Korea where Japanese B. mandarina occurs. The mitogenome presented here will provide useful information for evolutionary studies of the origin of the present day silkworm.
Figure 1. Phylogenetic tree of 16 B. mandarina strains available based on whole mitochondrial genome. The tree was built using Bayesian method with GTR + G + I model. Bootstrap values measured by the posterior probabilities are shown at the nodes. Rondotia menciana and Antheraea pernyi served as outgroups. The accession numbers are listed following silkworm name.
Disclosure statement
The authors declare no competing financial interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Astaurov BL, Rovinskaya IS. 1959. Chromosome complex of Ussuri geographical race of Bombyx mandarina M. with special reference to the problem of the origin of the domesticated silkworm, Bombyx mori. Cytology. 1:327–332.
- Hu XL, Cao GL, Xue RY, Zheng XJ, Zhang X, Duan HR, Gong CL. 2010. The complete mitogenome and phylogenetic analysis of Bombyx mandarina strain Qingzhou. Mol Biol Rep. 37:2599–2608.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Kong W, Yang J. 2015. The complete mitochondrial genome of Rondotia menciana (Lepidoptera: Bombycidae). J Insect Sci. 15:48.
- Li D, Guo Y, Shao H, Tellier LC, Wang J, Xiang ZH, Xia QY. 2010. Genetic diversity, molecular phylogeny and selection evidence of the silkworm mitochondria implicated by complete resequencing of 41 genomes. BMC Evol Biol. 10:81.
- Liu Y, Li Y, Pan M, Dai F, Zhu X, Lu C, Xiang Z. 2008. The complete mitochondrial genome of the Chinese oak silkmoth, Antheraea pernyi (Lepidoptera: Saturniidae). Acta Biochim Biophys Sin (Shanghai). 40:693–703.
- Pan MH, Yu QY, Xia YL, Dai FY, Liu YQ, Lu C, Zhang Z, Xiang ZH. 2008. Characterization of mitochondrial genome of Chinese wild mulberry silkworm, Bombyx mandarina (Lepidoptera: Bombycidae). Sci China Ser C-Life Sci. 51:693–701.
- Shimada T, Kurimoto Y, Kobayashi M. 1995. Phylogenetic relationship of silkmoths inferred from sequence data of the arylphorin gene. Mol Phylogenet Evol. 4:223–234.
- Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol Biol Evol. 19:1385–1389.
