Abstract
The complete mitochondrial genomes of two Sri Lankan junglefowl (Gallus lafayetti: CJF) individuals were sequenced by using next-generation sequencing technique. Samples were collected from Rathnapura and Pelmadulla areas in Sri Lanka. The complete mitochondrial DNA is 16,839 bp in length, with a typical mitogenome structure composed of a non-coding control region, 22 tRNA, two rRNA, and 13 protein-coding genes. Overall base composition is 30% A, 23.9% T, 32.3% C, and 13.6% G indicating high content of 54.0% A + T for both individuals. Phylogenetic analysis reveals that CJF samples cluster with the clade of the green junglefowl (Gallus varius) and red junglefowl (Gallus gallus) than to grey junglefowl (Gallus sonerattii: GyJF). This result can be subsequently used to provide essential information for junglefowl evolution.
Introduction
The Sri Lankan junglefowl (Gallus lafayetti: CJF), also known as Ceylon junglefowl, is one of the species in genus Gallus among the four species (grey junglefowl (G. sonneratii; GyJF), green junglefowl (G. varius; GJF), red junglefowl (G. gallus; RJF) and CJF) (Delacour Citation1977; Sibley and Ahlquist Citation1990; Johnsgard Citation1999). It is endemic to Sri Lanka and considered as the national bird of Sri Lanka, distributed in Yala National Park in dry zone, Sinharaja rain forest and most of tea estates. Being geographically isolated in island from Indian sub-continent, very few genomic and evolutionary studies have been carried out on CJF to understand its phylogeny. The total genomic DNA of the two CJF individuals from Rathnapura and Pelmadulla areas in Sri Lanka was extracted from the whole blood with standard phenol/chloroform methods. The PCR, library construction, next-generation sequencing, and de novo assembly for mitochondrial DNA (mtDNA) genomes followed the previous protocol (Chen et al. Citation2016). Caveats were followed for quality control in mtDNA data analyses (Shi et al. Citation2014). The variants were scored and checked manually relative to the reference sequence AP003321 (Nishibori et al. Citation2005) and the bam file was exported by Torrent Suite 5.0.2 to confirm the scored variants by using Integrative Genomics Viewer (Thorvaldsdóttir et al. Citation2013).
We obtained the complete mtDNA genomes of two CJF individuals (GSA No. PRJCA000289 and PRJCA000290) in our study. We described 16,839 bp of CJF mitochondrial genomic sequences, including a non-coding control region, 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and 13 protein-coding genes. On the average, overall base composition of the mitochondrial genomes is as follows: 30.1% A, 23.9% T, 32.3% C, and 13.6% G, showing high content of 54.0% A + T for both samples. Nucleotide composition was estimated by MEGA 7.0 (Kumar et al. Citation2016). The three CJF sequences (two de novo and one from previous study, Nishibori et al. Citation2005) had 100% bootstrap support from neighbour-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) analyses. Therefore, this provides further evidence for the validity of the sequences obtained in our study.
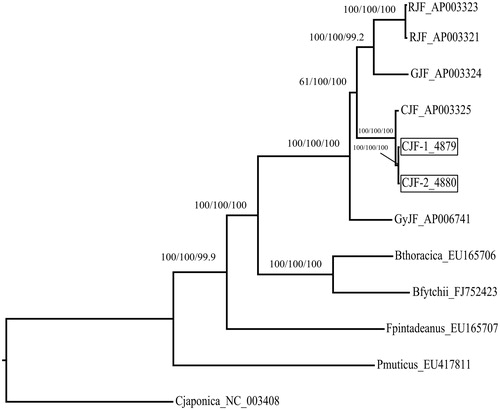
The phylogenetic position of CJF was estimated from complete mtDNA sequences. Neighbour-joining analyses were performed in MEGA 7.0, ML calculated in RaxML (Randomized A(x)ccelerated Maximum Likelihood) (Stamatakis et al. Citation2008; https://embnet.vital-it.ch/raxml-bb/), and MP estimated using PAUP* (V4.0) (Swofford Citation2003). The phylogenetic analysis results show that the Gallus four species clustered in three different clades. RJF and GJF grouped in closer clades while CJF is more distance from GyJF. This is in contrast with Helm-Bychowski and Wilson (Citation1986) and Fumihito et al. (Citation1996), our findings showed that having a common ancestor CJF is more closely related to both GJF and RJF than to GyJF which is in accordance with Guan et al. (Citation2009). This study will contribute to the phylogenetic analyses in Galliformes ().
Figure 1. Phylogenetic tree of the relationships among Galliformes based on mtDNA genome sequences. Pavo muticus served as the outgroup. Numbers above each node indicates the NJ, ML, and MP bootstrap support values, respectively. All the species’ accession numbers are listed as follows: Gallus gallus (RJF) AP003321, AP003323; G. varius (GJF) AP003324; G. sonneratii (GyJF) AP006741; G. lafayettei (CJF) AP003325, PRJCA000289–4880, PRJCA000290–4879; Bambusicola thoracicus (Bthoracica) EU165706; B. fytchii (Bfytchii) FJ752423; Coturnix japonica (Cjaponica) NC_003408; Francolinus pintadeanus (Fpintadeanus) EU165707; Pavo muticus (Pmuticus) EU417811.
Acknowledgements
We thank Director General and other staff of Dept. of Wild Life Conservation, Sri Lanka, for their assistance in sampling.
Disclosure statement
The authors have declared that no competing interests exist and are solely responsible for this paper.
Additional information
Funding
References
- Chen X, Ni G, He K, Ding ZL, Li GM, Adeola AC, Murphy RW, Wang WZ, Zhang YP. 2016. An improved de novo pipeline for enrichment of high diversity mitochondrial genomes from Amphibia to high-throughput sequencing. bioRxiv. https://doi.org/10.1101/080689.
- Delacour J. 1977. The pheasants of the world. 2nd ed. Hindhead (UK): World Pheasant Association and Spur Publications.
- Fumihito A, Miyake T, Takada M, Shigu R, Endo T, Gojobori T, Kondo N, Ohno S. 1996. Monophyletic origin and unique dispersal patters of domestic fowls. Proc Natl Acad Sci USA. 93:6792–6795.
- Guan XJ, Silva P, Gyenai BK, Xu J, Geng T, Tu Z, Samuels CD, Smith EJ. 2009. The mitochondrial genome sequence and molecular phylogeny of the Turkey, Meleagris gallopava. Anim Genet. 40:134–141.
- Helm-Bychowski KM, Wilson AC. 1986. Rates of nuclear DNA evolution in pheasant-like birds: evidence from restriction maps. Proc Natl Acad Sci USA. 83:688–692.
- Johnsgard PA. 1999. The pheasants of the world; biology and natural history. 2nd ed. Washington (DC): Smithsonian Institution Press.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Nishibori M, Shimogiri T, Hayashi T, Yasue H. 2005. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim Genet. 36:367–375.
- Shi NN, Fan L, Yao YG, Peng MS, Zhang YP. 2014. Mitochondrial genomes of domestic animals need scrutiny. Mol Ecol. 23:5393–5397.
- Sibley CG, Ahlquist JE. 1990. Phylogeny and classification of birds: a study in molecular evolution. New Haven (CT): Yale University Press.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 75:758–771.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates.
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14:178–192.
