Abstract
The nearly complete mitochondrial genome of Paederus fuscipes (GenBank accession no. MG581161) is 17,644 bp in size, containing 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and a partial control region. The gene order is similar to the typical insect mitochondrial genome. Maximum likelihood tree recovered the monophyly of Staphylininae, Pselaphinae, Paederinae and Aleocharinae. Additionally, Staphylininae is a sister group to Paederinae.
The family Staphylinidae, belonging to Coleoptera, contains over 61,000 described extant species distributed worldwide (Newton Citation2015). We determined the nearly complete mitochondrial genomes of Paederus fuscipes to expand the data of mitochondrial genomes of Staphylinidae and to better understand the phylogenetic relationships of Staphylinidae.
Paederus fuscipes was collected in Zhengzhou, China (the geospatial coordinates: 113.635°E, 34.723°N), the primary specimen can be obtained by the Entomological Museum of Henan Agricultural University (voucher no. MT-Zz15091901). Total genomic DNA was extracted from muscular tissue preserved in the absolute ethyl alcohol at −20 °C using the TIANamp Micro DNA Kit (Tiangen Biotech Co., Ltd, Beijing, China). We reconstructed the nearly complete mitochondrial genome of P. fuscipes from the library composed of the Genomic DNA of P. fuscipes and other unrelated insects, by using sequencing of PE 150 in the Illumina HiSeq 2500 platform. The raw reads were de novo assembled by the SOAPdenovo software (Zhao et al. Citation2011) with an average 358.98 × coverage. We identified the nearly complete mitochondrial genome of P. fuscipes from a single large contig of 17,386 bp by blasting the mitochondrial rrnS gene bait sequence against the assembled contigs with BioEdit7.0.9.0 (Hall Citation1999).
The nearly complete mitochondrial genomes of P. fuscipes (GenBank accession no. MG581161) is 17,644 bp in length, contains 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and a partial control region. The gene arrangement is similar to the typical insect mitochondrial genomes (Wolstenholme Citation1992). All protein-coding genes of P. fuscipes start with the start codon ATN except for nad1, which begins with putative start codon TTG. Ten protein-coding genes of P. fuscipes use TAA or TAG as stop codon, but three of the genes (i.e. cox2, cox3 and nad4) use T as incomplete stop codon. All 22 tRNA genes can be folded into the typical cloverleaf structure, but the anticodon of trnS1 is UCU rather than usual GCU. Although the control region has a length of 3119 bp, it is still incomplete due to the lack of the overlapping sequences to connect two ends of this region.
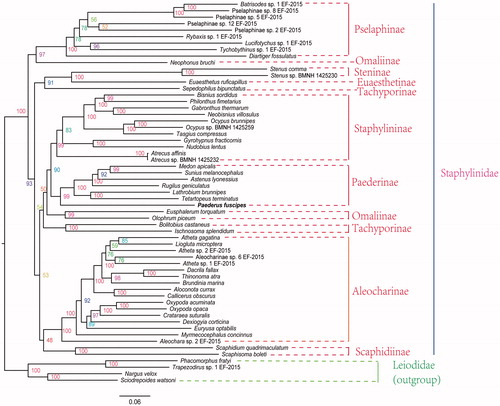
Phylogenetic analysis shows that Staphylininae, Pselaphinae, Paederinae and Aleocharinae are monophyletic (Brunkea et al. Citation2015; Mckenna et al. Citation2015) (). Staphylininae is a sister group to Paederinae ( Crowson Citation1955; Lawrence and Newton Citation1995; Beutel and Molenda Citation1997; Assing Citation2000; Solodovnikov and Newton Citation2005; Solodovnikov et al. Citation2013; Mckenna et al. Citation2015). However, Omaliinae and Tachyporinae are non-monophyletic (Mckenna et al. Citation2015).
Figure 1. Maximum-likelihood tree of the mitogenomes of P. fuscipes and 58 other species from GenBank. The maximum-likelihood analysis was reconstructed by concatenated nucleotide sequences of 13 mitochondrial protein genes (10,869 bp) using IQ-TREE (Nguyen et al. Citation2015). Numbers alongside nodes refer to bootstrap support values. The name in the rough line is our determined mitochondrial genome. All 59 species accession numbers are listed as below: Eusphalerum torquatum (KT780648), Neophonus bruchi (KT780663), Olophrum piceum (NC_028605), Pselaphinae sp. 5 EF-2015 (KT780684), Pselaphinae sp. 8 EF-2015 (KT780689), Pselaphinae sp. 12 EF-2015 (KT780678), Batrisodes sp. 1 EF-2015 (KT780631), Diartiger fossulatus (KT780644), Lucifotychus sp. 1 EF-2015 (KT780656), Rybaxis sp. 1 EF-2015 (KT780672), Tychobythinus sp. 1 EF-2015 (KT780701), Pselaphinae sp. 2 EF-2015 (KT780680), Scaphidium quadrimaculatum (NC_028609), Scaphisoma boleti (KT780674), Astenus lyonessius (KT780626), Lathrobium brunnipes (KT780654), P. fuscipes (MG581161), Medon apicalis (KT780658), Rugilus geniculatus (NC_028608), Sunius melanocephalus (KT780696), Tetartopeus terminatus (NC_028613), Euaesthetus ruficapillus (KT780646), Stenus sp. BMNH 1425230 (KT876913), Stenus comma (KT780694), Atrecus sp. BMNH 1425232 (KT876882), Atrecus affinis (NC_028597), Neobisnius villosulus (KT780662), Philonthus fimetarius (KT780669), Bisnius sordidus (KT780632), Gabronthus thermarum (NC_028601), Ocypus sp. BMNH 1425259 (KT876908), Ocypus brunnipes (KT780665), Tasgius compressus (KT780697), Gyrohypnus fracticornis (KT780650), Nudobius lentus (KT780664), Atheta gagatina (KT780629), Aleochara sp. 2 EF-2015 (KT780622), Aleocharinae sp. 6 EF-2015 (KT780687), Aloconota currax (KT780624), Atheta sp. 2 EF-2015 (KT780628), Atheta sp. 1 EF-2015 (KT780627), Callicerus obscurus (NC_028598), Liogluta microptera (NC_028602), Euryusa optabilis (NC_028600), Myrmecocephalus concinnus (NC_028604), Dexiogyia corticina (KT780643), Oxypoda acuminata (NC_028606), Crataraea suturalis (KT780639), Oxypoda opaca (JX412751), Brundinia marina (KT780635), Thinonoma atra (KT780699), Dacrila fallax (NC_028599), Bolitobius castaneus (KT780633), Ischnosoma splendidum (KT780653), Sepedophilus bipunctatus (NC_028611), Nargus velox (KT780661), Trapezodirus sp. 1 EF-2015 (KT780700), Phacomorphus fratyi (NC_028607), Sciodrepoides watsoni (NC_028610).
Disclosure statement
All authors have read and approved the final manuscript. No conflict of interest was reported by the authors.
Additional information
Funding
References
- Assing V. 2000. A taxonomic and phylogenetic revision of Maorothiini trib. n. from the New Zealand subregion (Coleoptera: Staphylinidae, Staphylininae). Beitr Entomol. 50:3–64.
- Beutel RG, Molenda R. 1997. Comparative morphology of selected larvae of Staphylinoidea (Coleoptera, Polyphaga) with phylogenetic implications. Zoologischer Anzeiger. 236:37–67.
- Brunkea AJ, Chatzimanolis S, Schillhammer H, Solodovnikov A. 2015. Early evolution of the hyperdiverse rove beetle tribe Staphylinini (Coleoptera: Staphylinidae: Staphylininae) and a revision of its higher classification. Cladistics. 32:427–451.
- Crowson RA. 1955. The natural classification of the families of Coleoptera. London: Nathaniel Lloyd.
- Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Lawrence JF, Newton AF, Jr. 1995. Families and subfamilies of Coleoptera (with selected genera, notes, references and data on family-group names). In: Pakaluk J, Slipinski SA, editors. Biology, Phylogeny and Classification of Coleoptera: Papers Celebrating the 80th Birthday of Roy A. Crowson. Warsaw: Muzeumí Instytut Zoologii PAN; p. 779–1006.
- Mckenna DD, Farrell BD, Caterino MS, Farnum CW, Hawks DC, Maddison DR, Seago AE, Short AEZ, Newton AF, Thayer MK. 2015. Phylogeny and evolution of Staphyliniformia and Scarabaeiformia: forest litter as a stepping stone for diversification of nonphytophagous beetles. Syst Entomol. 40:35–60.
- Newton AF. 2015. Beetles (Coleoptera) of Peru: a survey of the families. Staphylinidae Latreille, 1802. J Kansas Entomol Soc. 88:283–304.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Solodovnikov AY, Newton AF. 2005. Phylogenetic placement of Arrowinini trib. n. within the subfamily Staphylininae (Coleoptera: Staphylinidae), with revision of the relict South African genus Arrowinus and description of its larva. Syst Entomol. 30:398–441.
- Solodovnikov A, Yue YL, Tarasov S, Ren D. 2013. Extinct and extant rove beetles meet in the matrix: early Cretaceous fossils shed light on the evolution of a hyperdiverse insect lineage (Coleoptera: Staphylinidae: Staphylininae). Cladistics. 29:360–403.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
