Abstract
Plagiorhegma dubia Maxim. is a traditional Chinese medicinal herb from Plagiorhegma, Berberidaceae, which is distributed in the northeast of China, Korea, Russia. The complete chloroplast genome is 152,468 bp in length, with large single copy (LSC 82,257 bp) and small single copy (SSC 16,599 bp) regions separated by a pair of inverted repeats (IR 26 805 bp). The genome has a total of 113 genes including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Phylogenetic analysis shows that P. dubia is closely related with Sinopodophyllum hexandrum and Epimedium species. The results are of great implication for the development and utilization of P. dubia and the phylogenetic researches on Berberidaceae.
Plagiorhegma dubia Maxim is a perennial herbaceous plant and the only species of the genus Plagiorhegma of Berberidaceae, Ranales, which is distributed in Northeast China, Korea, Russia (Ying et al. Citation2011). As the traditional Chinese medicinal material, the rhizome of P. dubia contains berberine, and has antipyretic, antidotal, stomachic and antidiarrhoeal clinical effects (Arens et al. Citation1985; Jeong and Sivanesan Citation2016). Furthermore, P. dubia has potential as a new ornamental garden plant which has attractive heart-shaped leaves and light purple flowers. The species has potential as a new ornamental garden plant (Rhie et al. Citation2014).
The total genomic DNA of P. dubia was extracted from its fresh leaves which were collected in Tonghua of Jinlin Province, China (N 41°43′48″ E 125°59′24″). An Illumina paired-end (PE) genomic library was prepared and sequenced. High quality reads were obtained from the total pair-end (PE) raw reads and assembled by the CLC-quality genome assembler (ver 4.06bata) (Kim et al. Citation2015). All of the contig sequences were aligned and ordered to the reference cp genome of Nandina domestica (NC_DQ923117) using MUMmer (Kurtz et al. Citation2004). Sequence gaps were filled by Gapcloser included in the SOAP package vl.12 (Li et al. Citation2010). Four junctions between LSC/IRs and SSC/IRs were validated with PCR-based conventional Sanger sequencing.
The genes in the chloroplast genome were predicted by the Dul Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004). The BLAST tools and ORF finder at NCBI website (http://www.ncbi.nlm.nih.gov/) were also used in the gene annotations. The tRNA genes were verified with tRNAscan-SE (Lowe and Eddy Citation1997). The circular cp genome maps were drawn by the Organellar Genome DRAW tool (OGDRAW; Lohse et al. Citation2007) with subsequent manual editing.
The complete chloroplast genome of P. dubia was submitted to NCBI, and the accession number of nucleotide sequence is MG397139. The nucleotide sequence was 152,468 bp, and was assembled into a single circular which presented a typical quadripartite structure including one large single-copy region (82,257 bp), one small single copy (16,599 bp), and a pair of inverted repeat regions (IRa and IRb) of 26805 bp. The GC content of the chloroplast genomes was 38.15%. The 113 unique genes in the sequence were composed of 79 protein-coding genes, 30 tRNA genes and four rRNA genes.
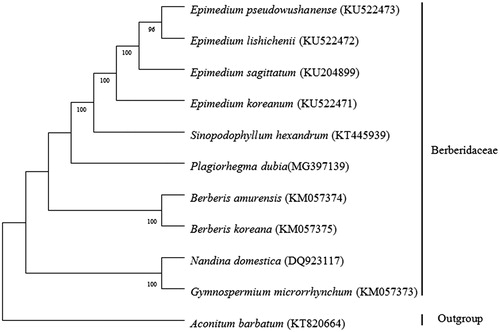
The phylogenetic analyses sampled P. dubia and other nine Berberdaceae species. These nine species had been sequenced on chloroplast genome, including four Epimedium species (Lee et al. Citation2015; Sun et al. Citation2016; Zhang et al. Citation2016), Sinopodophyllum hexandrum (Meng et al. Citation2017), two Berberis species (NC_KM057374, NC_KM057375), Nandina domestica (Moore et al. Citation2006), Gymnosepermium microrrhynchum (NC_KM057373). ML phylogenetic tree was constructed based on the entire chloroplast protein-coding sequences of these 11 species using MEGA7.0 (Kumar et al. Citation2016). The results showed that the ten Berberidaceae species grouped into a monophyletic branch (). In accordance with previous phylogenetic studies on Berberidaceae (Kim and Jansen Citation1994; Wang et al. Citation2007; Zhang et al. Citation2012), inter-genera relationships of the family were closely related with chromosome base number. N. domestica with x = 10 and G. microrrhynchum with x = 8 clustered into a clade. Berberis amurensis and B. korena are with x = 7 and formed a branch. Epimedium koreanum, E. sagittatum, E. lishichenii, E. pseudowushanense, S. hexandrum, P. dubia are with x = 6 and had most closest relationships. The results are of great implication for the Phylogenetic researches on Berberidaceae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Arens H, Fischer H, Leyck S, Römer A, Ulbrich B. 1985. Antiinflammatory compounds from Plagiorhegma dubium cell culture. Planta Medica. 51:52–56.
- Jeong BR, Sivanesan I. 2016. Micropropagation, berberine content and antitumor activity of Jeffersonia dubia (Maxim.) Benth et Hook. Plant Cell Tissue Organ Culture. 124:453–458.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45s nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Kim YD, Jansen RK. 1994. Characterization and phylogenetic distribution of a chloroplast DNA rearrangement in the Berberidaceae. Plant System Evol. 193:107–114.
- Kumar S, Stecher G, Tamura K. 2016. Mega 7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
- Lee JH, Kim K, Kim NR, Lee SC, Yang TJ, Kim YD. 2015. The complete chloroplast genome of a medicinal plant Epimedium koreanum Nakai (Berberidaceae). Mitochondrial DNA Part A. 27:4342–4343.
- Li RQ, Zhu HM, Ruan J, Qian WB, Fang XD, Shi ZB, Li Y, Li S, Shan G, Kristiansen K, et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Research. 20:265–272.
- Lohse M, Drechsel O, Bock R. 2007. Organellar Genome DRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics. 52:267–274.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Meng LH, Liu RJ, Chen JB, Ding CX. 2017. The complete chloroplast genome of Sinopodophyllum hexandrum Ying (Berberidaceae). Mitochondrial DNA Part A. 28:342–343.
- Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, Folta KM, Soltis DE. 2006. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 6:17.
- Rhie YH, Lee SY, Jung HH, Kim KS. 2014. Light intensity influences photosynthesis and crop characteristics of Jeffersonia dubia. Korean J Horticult Sci Technol. 32:584–589.
- Sun YX, Moore MJ, Zhang S, Soltis PS, Soltis DE, Zhao T, Meng A, Li X, Li J, Wang H. 2016. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of ir gene content evolution. Molecular Phylogenetics & Evolution. 96:93–101.
- Wang W, Chen ZD, Liu Y, Li RQ, Li JH. 2007. Phylogenetic and biogeographic diversification of Berberidaceae in the northern hemisphere. Systematic Bot. 32:731–742.
- Wyman SK, Jansen RK, Boor JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Ying TS, Boufford DE, Brach AR. 2011. “Berberidaceae”, in Flora of China. Wu ZY, Raven PH, Hong DY, editors. Beijing: Science Press/St. Louis (MO): Missouri Botanical Garden Press; p. 714–801.
- Zhang MY, Lu L, Li DZ, Wang H. 2012. Evolution of the pollen in the family Berberidaceae. Plant Diversity Resour. 34:1–11.
- Zhang YJ, Du LW, Liu A, Chen JJ, Wu L, Hu WM, Zhang W, Kim K, Lee SC, Yang TJ. 2016. The complete chloroplast genome sequences of five Epimedium Species: lights into phylogenetic and taxonomic analyses. Frontiers Plant Sci. 7:306.