Abstract
The Hycleus cichorii and Hycleus phaleratus are two species of medicinal meloids widely distributed in southwest of China. We sequenced an annotated the complete mitochondrial genomes of H. cichorii and H. phaleratus, and the mitogenomes are 15,847 and 16,004 bp in length, respectively. Every mitochondrial genome encodes 13 proteins, 2 ribosomal RNAs, 22 tRNAs, and a control region with the identical arrangement to other beetles. The preliminary phylogenetic analysis with mitochondrial genomes of nine meloid species further confirmed the status of these two species.
Blister beetles have a long history in human medicine for its irritant property (Bologna Citation1991; Tan et al. Citation1995). Their released oils contain the skin irritant cantharidin, which is a defensive terpenoid and sexual pheromone. The mitochondrial genomes of meloids were described and utilized to infer their phylogenetic implications (Du et al. Citation2016; Yuan et al. Citation2016; Du et al. Citation2017). However, some relationships among this family were unresolved due to the limitation of available mitochondrial information. In this study, we present the complete mitochondrial genomes of H. cichorii and H. phaleratus, which would be a significant increase in further study of mitochondrial genome architecture and meloid phylogenetics. Specimens were collected in Luodian, Guizhou, China, and disposed in Institute of Entomology/Special Key Laboratory for Development and Utilization of Insect Resources, Guizhou University. The high-throughput sequencing method was used to obtain the complete mitochondrial genomes.
The complete mitochondrial genome of H. cichorii (accession no. MF491388) and H. phaleratus (accession no. MF491389) are 15,847 and 16,004 bp in length, respectively. Every mitochondrial genome encodes the typical 37 genes including 13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs. The A + T contents of two mitochondrial genomes are 73.0% and 69.9%, respectively. All 13 protein-coding genes used typical ATN start codons, including eight Met (ATA & ATG) and five Ile (ATT). Conventional stop codons could be assigned to most of the PCGs, while cox1, cox2, nad5, and nad4 terminated with incomplete stop codon T. All tRNAs could be folded into the typical clover-leaf structure, except trnS(AGN) lacked a dihydrouridine (DHU) arm, which was replaced by a simple loop. The lengths of rrnL were determined to be 1,281 and 1,276 bp in H. phaleratus and H. cichorii respectively, and the lengths of rrnS were 793 bp in both species. The 1,129 and 1,128 bp control regions of H. phaleratus and H. cichorii mitogenome were located between rrnS and trnI.
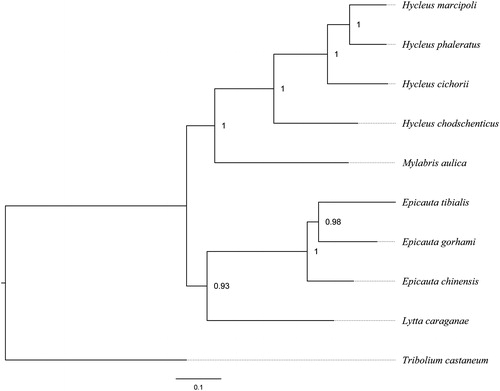
Phylogenetic analysis was performed based on 13 protein-coding genes of nine meloid species, by using Bayesian Inference. The result further confirmed that H. phaleratus and H. cichorii were closely related to H. marcipoli and H. chodschenticus, and all Hycleus species were recovered as a monophyly to sister to M. aulica (). Besides, the relationships among Meloidae were congruent with the previous study (Du et al. Citation2017).
Figure 1. The Bayesian phylogenetic tree of nine meloids was constructed using the dataset of 13 protein-coding genes, with the tenebrionoid Tribolium castaneum employed as the outgroup. The numbers abutting nodes refer to Bayesian posterior probabilities. Sequence data used in this study are the following: Hycleus marcipoli (KX161857), Hycleus chodschenticus (KT808466), Mylabris aulica (KX161860), Epicauta chinensis (KP692789), Epicauta tibialis (KX161855), Epicauta gorhami (KX161854), Lytta caraganae (KX161859), Tribolium confusum (NC_026702).
Acknowledgements
This work was supported by the National Natural Science Foundation of China [No. 81460576].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bologna MA. 1991. Fauna de Italia. XXVIII. Coleoptera Meloidae. Bologna: Edizioni Calderini.
- Du C, He S, Song X, Liao Q, Zhang X, Yue B. 2016. The complete mitochondrial genome of Epicauta chinensis (Coleoptera: Meloidae) and phylogenetic analysis among coleopteran insects. Gene. 578:274–280.
- Du C, Zhang L, Lu T, Ma J, Zeng C, Yue B, Zhang X. 2017. Mitochondrial genomes of blister beetles (Coleoptera: Meloidae) and two large intergenic spacers in Hycleus genera. Bmc Genomics. 18:698.
- Tan J, Zhang Y, Wang S, Deng Z, Zhu C. 1995. Investigation on the natural resources and utilization of the Chinese medicinal beebles-Meloidae. Acta Entomologica Sinica. 38:324–331.
- Yuan ML, Zhang QL, Zhang L, Guo ZL, Liu YJ, Shen YY, Shao R. 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol Phylogenet Evol. 104:99–111.
