Abstract
The complete chloroplast genome sequence of Berchemia berchemiifolia, rare plant to Korea, was determined in this study. The total genome size was 160,410 bp in length, containing a pair of inverted repeats (IRs) of 26,514 bp, which were separated by a large single copy (LSC) and small single copy (SSC) of 88,627 bp and 18,755 bp, respectively. The overall GC contents of the chloroplast genome were 37.2%. One hundred twenty nine genes were annotated, including 84 protein coding genes, 37 tRNA genes, and 8 rRNA genes. In these genes, 18 genes contained one or two introns. The phylogenetic tree showed that Berchemia berchemiifolia was most closely related to Berchemiella wilsonii.
Genus Berchemia, belonging to Rhamnaceae, include about 32 taxa and distributed mainly in temperate and tropical regions of East to Southeast Asia (Michaux and Hedwig Citation2007). Among the Berchemia species, Berchemia berchemiifolia is distributed only in the Korea peninsula and Japan. In the Korea, especially, this species is distributed only in limited regions in Kyeongsangbuk-do and Chungcheongbuk-do Province, and three natural habitats in Chungcheongbuk-do Province are designated as natural monuments and protected. According to these reasons, it is listed in the rare plant in Korea as a ‘Vulnerable (VU)’ grade by IUCN category (Korea National Arboretum Citation2009).
In this study, we reported the complete chloroplast genome sequence of B. berchemiifolia, and this data will be used as an important information for protection strategy of species and the study of genome diversity. The plant materials were sampled from Sadam-ri (Goesan-gun, Chungcheongbuk-do Province, Korea). The voucher specimen has been deposited at the Kangwon National University Herbarium (KWNU93462). Total DNA was extracted using DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA). The genomic DNA was sequenced using the Miseq (Illumina Inc., San Diego, CA, USA). Using these sequence data, we assembled the complete chloroplast genome with Geneious 7.1.9 (Biomatters Ltd, New Zealand). Annotation was performed with Dual Organellar GenoMe Annotator (DOGMA) using default parameters to predict protein-coding genes, transfer RNA (tRNA) genes and ribosomal RNA (rRNA) genes (Wyman et al. Citation2004). BLASTX was used to further identify positions of genes with intron by searching against published plastid genome database. Genome map was drawn by OGDraw v 1.2 (Lohse et al. Citation2007). The complete cp genome sequence of B. berchemiifolia was submitted to GenBank under accession number MG739656.
The chloroplast genome of B. berchemiifolia was 160,410 bp in length with 37.2% GC content and composed of large single copy (LSC) region of 88,627 bp, small single copy (SSC) region of 18,755 bp, and two inverted repeat (IR) regions 26,514 bp. The gene content and order of plastid genome were almost identical to Rhamnaceae species (Ma et al. Citation2017; Wang et al. Citation2017) such as Berchemiella wilsonii (GenBank accession KY926621), Ziziphus jujuba (GenBank accession KU351660), and Z. jujuba var. spinosa (GenBank accession KX26630). A total of 129 genes were annotated, including 84 protein coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. The rps12 is trans-spliced gene with the 5′ exon located in the LSC region and 3′ exon was duplicated in the IR regions. Eighteen genes contain one or two introns (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, petB, petD, atpF, ndhA, ndhB, clpP, rpl2, rpl16, rps12, rps16, rpoC1, and ycf3).
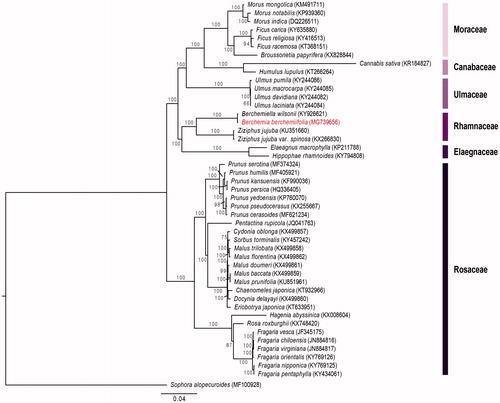
To construct the phylogenetic tree, a total of 65 protein coding genes of 45 Rosales species and one outgroup (Sophora alopecuroides) were compiled into a single file of 74,041 bp and aligned with MAFFT (Katoh et al. Citation2002) and then, we conducted maximum likelihood (ML) analysis using RAxML v.7.4.2 with 1000 bootstrap replicates and GTR + I model (Stamatakis Citation2006). The phylogenetic tree () showed that Rhamnaceae are monophyletic and sister to Elaegnaceae. Also, B. berchemiifolia was most closely related to Berchemiella wilsonii with high support (BS = 100).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Korea National Arboretum. 2009. Rare Plants Data Book in Korea. Pocheon: Korea National Arboretum.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Ma Q, Li S, Bi C, Hao Z, Sun C, Ye N. 2017. Complete chloroplast genome sequence of a major economic species, Ziziphus jujuba (Rhamnaceae). Curr Genet. 63:117–129.
- Michaux O, Hedwig R. 2007. Flora of China. 12:124–130.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wang YH, Chen SY, Zhang SD. 2017. Characterization of the complete chloroplast genome of Berchemiella wilsonii var. wilsonii (Rhamnaceae), an Endangered species endemic to China. Conserv Genet Resour. First Online 1–3.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.