Abstract
Spring orchid (Cymbidium goeringii) is one of the most important species belonging to Orchidaceae owing to its aesthetic appeal, fragrant flowers and ideal characteristics for using as a houseplant. In this study, the complete chloroplast genome of Korean C. goeringii acc. smg222 was determined by Illumina sequencing. The circular double-stranded DNA of 148,441 bp consisted of two inverted repeat regions of 25,610 bp each, a large single copy region of 83,311 bp, and a small single copy region of 13,910 bp. The genome contained 122 genes, of which 104 were unique and 18 were duplicated within the IRs. The 104 unique genes included 70 protein-coding genes, 30 distinct tRNA genes, and four rRNA genes. Phylogenetic tree analysis revealed that C. goeringii acc. smg222 was clustered with Cymbidium kanran, a cymbidium species native to Korea.
Cymbidium orchids (Orchidaceae) are among the most popular orchids worldwide and it is distributed in tropical, subtropical Asia and northeastern Australia (Yang et al. Citation2013). Spring orchid (C. goeringii) is one of the most important species belonging to Cymbidium (Wang et al. Citation2009) owing to its aesthetic appeal and fragrant flowers for using as a decorative pot flowers (Liu et al. Citation2014). During the long cultivation history, most of the commercially important and attractive cymbidiums have been produced by selection on somatic mutations during vegetative propagation and hybrids derived from other species (Wang et al. Citation2009; Liu et al. Citation2014). These currently recognized variants are often complex, involving several species in their ancestry (Obara-Okeyo and Kako Citation1998). Chloroplasts, as one of the essential organelles, have become an effective method for phylogenetic analysis (Parks et al. Citation2009). Here, we report the complete chloroplast genome of C. goeringii acc. smg222, a spring orchid native to Korea, and generated phylogenetic tree to provide an insight into genetic relationships of Cymbidium and related species.
Total genomic DNAs were extracted from mature leaves obtained from the Saemangeum Bio Center, Republic of Korea (35°56′31.2″N, 126°48′53.0″E). An Illumina paired-end DNA library (average insert size of 500 bp) was constructed and sequenced by Illumina HiSeq platform. Totally, 2.2 Gb paired-end reads were obtained and assembled using the CLC Genome Assembler (version beta 4.6; CLC Bio, Aarhus, Denmark) with a minimum overlap size of 200 bp and maximum bubble size of 50 bp for the de Bruijn graph. Chloroplast contigs were selected from the initial assembly by performing a BLAST (version 2.2.31) search against the reference chloroplast genome of C. kanran (GenBank accession: KU179435) using CLC. The complete chloroplast genome was annotated and assembled with Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004) and CpGAVAS (Liu et al. Citation2012). The whole chloroplast genome sequence has been deposited to NCBI GenBank with the accession number of MF421552.
The C. goeringii acc. smg222 chloroplast genome was composed of a single, circular double-stranded DNA molecule, consisting of a pair of IRs (25,610 bp) separated by the LSC (83,311 bp) and SSC (13,910 bp) regions. The overall GC content was 37.1%. The chloroplast genome contains 122 genes, of which 104 were unique and 18 were duplicated in the IR regions. The 104 unique genes included 70 protein-coding, 30 transfer RNA, and four ribosomal RNA genes.
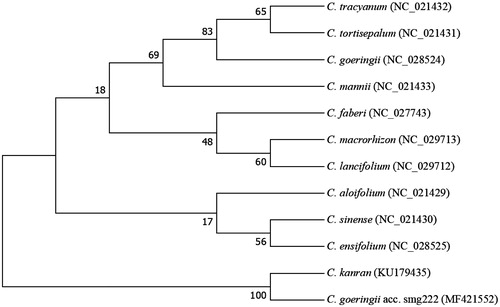
We also constructed a phylogenetic analysis with the complete chloroplast genome of C. goeringii acc. smg222 and 11 other species from the family of Cymbidium (). The neighbour-joining phylogenetic tree analysis was performed using MEGA 7.0 (Kumar et al. Citation2016). The tree showed that C. goeringii acc. smg222 was subgrouped with C. kanran, a Korean landrace (Chung et al. Citation1985). The newly characterized C. goeringii acc. smg222 chloroplast genome reported here provides a useful tool not only for studying of the evolutionary history of Cymbidium, but also for population structure and phytogeographic studies for Cymbidium.
Disclosure statement
No conflict of interest was reported by the authors.
Additional information
Funding
References
- Chung JD, Chun CK, Kim SS, Lee JS. 1985. Factors affecting growth of rhizome and organogenesis of Korean native Cymbidium kanran. J Kor Soc Hortic Sci (Korea R.). 26:281–288.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Liu X, Huang Y, Li F, Xu C, Chen K. 2014. Genetic diversity of 129 spring orchid (Cymbidium goeringii) cultivars and its relationship to horticultural types as assessed by EST-SSR markers. Sci Hortic. 174:178–184.
- Obara-Okeyo P, Kako S. 1998. Genetic diversity and identification of Cymbidium cultivars as measured by random amplified polymorphic DNA (RAPD) markers. Euphytica. 99:95–101.
- Parks M, Cronn R, Liston A. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 7:84.
- Wang HZ, Wu ZX, Lu JJ, Shi NN, Zhao Y, Zhang ZT, Liu JJ. 2009. Molecular diversity and relationships among Cymbidium goeringii cultivars based on inter-simple sequence repeat (ISSR) markers. Genetica. 136:391–399.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang JB, Tang M, Li HT, Zhang ZR, Li DZ. 2013. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 13:84.