Abstract
The complete chloroplast genome sequence of Phedimus kamtschaticus, which commonly occurs in northeastern Asia was determined. The genome size was 151,652 bp, composed of one pair of inverted repeats (IRs) of 25,977 bp, which were separated by one large single-copy (LSC; 83,010 bp) and one small single-copy (SSC; 16,688 bp) region. The chloroplast genome contained 132 genes, including 88 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content was 37.8%. Phylogenetic analysis of the complete chloroplast genome suggested that P. kamtschaticus was most closely related to Ulleung Island insular endemic P. takesimensis.
The family Crassulaceae is a morphologically diverse and taxonomically complex angiosperm family, comprising 35 genera and approximately 1500 species (Berger Citation1930). While the Crassulaceae are considered a natural group, infrafamilial classification has been extremely difficult due to homoplasious morphological features (van Ham Citation1995; Soltis et al. Citation2000). The genus Phedimus (ca. 20 species in Asia and Europe) represents one lineage of nonmonophyletic subfamily Sedoideae. Traditionally, species of Phedimus have been treated as members of Sedum, but recent phylogenetic study strongly supported the monophyly of Phedimus and its segregation from Sedum (Mayuzumi and Ohba Citation2004). Of ca. 20 species of Phedimus, we selected P. kamtschaticus, which occurs commonly in the northeastern Asia, and sequenced its complete chloroplast genome because it is considered to be a continental progenitor of Ulleungdo and Dokdo Island endemic P. takesimensis in Korea.
We collected fresh leaves of P. kamtschaticus from Mt. Hambaek, Korea (GPS coordinates 37°09′16.6″N 128°54′53.4″E; altitude 1334 m above the sea level) and a voucher specimen (SKK014113) was deposited at the herbarium of Sungkyunkwan University (SKK). Total DNA was iso lated using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA), following the instructions of the manufacturer. Sequencing was done using the Illumina Miseq (Illumina Inc., San Diego, CA) platform and assembled by SPAdes 3.6.1 (Bankevich et al. Citation2012) and CLC Genomics Workbench v.5.5.1 (CLC Bio, Aarhus, Denmark). The chloroplast genome of P. kamtschaticus was annotated using the Dual Organellar GenoMe Annotator (DOGMA) tool (Wyman et al. Citation2004) and CpGAVAS (Liu et al. Citation2012) with plastid/bacterial genetic code. The tRNAs were confirmed using tRNAscan-SE with default settings (Schattner et al. Citation2005). Gene annotation was done using BLAST X, Geneious v.8.1.6. (Biomatters Ltd., Auckland, New Zealand), and then manually corrected for intron/exon boundaries. The complete chloroplast genome of P. kamtschaticus was aligned with 11 representative species of Saxifragales using MAFFT v.7 (Katoh and Standley Citation2013). Maximum likelihood (ML) analysis was conducted based on 13 complete chloroplast genomes (1 outgroup and 12 ingroup) using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015) with 1000 bootstrap (BS) replications.
Here, we reported the complete chloroplast genome sequence of P. kamtschaticus (MG680403), which has a total length of 151,652 bp with 37.8% GC content, and contained two inverted repeat regions (IRa and IRb) of 25,977 bp, separating a large single-copy (LSC) region of 83,010 bp and a small single-copy (SSC) region of 16,688 bp. The chloroplast genome contained 132 genes, including 88 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The structure, gene content and order, and GC content of P. kamtschaticus were similar to those of P. takesimensis chloroplast genome.
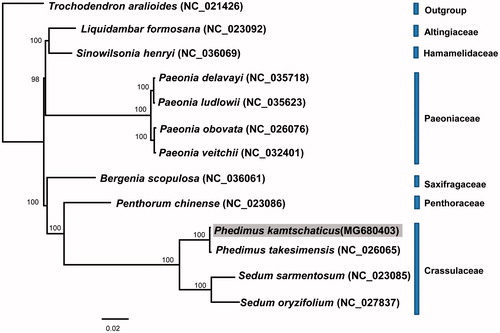
The phylogenetic relationship of P. kamtschaticus was determined based on 11 other representative chloroplast genomes in Saxifragales. The phylogenetic analysis showed that the family Crassulaceae, including four species of Phedimus and Sedum, was monophyletic (100% BS) (). P. kamtschaticus was most closely related to Ulleung Island endemic P. takesimensis in Korea, establishing a continental progenitor and insular derivative relationship.
Acknowledgements
We thank Seon A Yun, Hee-Young Gil, and Joon-Mo Lee for technical help during the assembly of chloroplast genome.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Berger A. 1930. Crassulaceae. In: Engler A, Prantl K, editors. Die Natürlichen Pflanzenfamilien, Vol. 2, 18a. Leipzigp: W. Engelmann; p. 352–483.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Mayuzumi S, Ohba H. 2004. The phylogenetic position of eastern Asian Sedoideae (Crassulaceae) inferred from chloroplast and nuclear DNA sequences. Syst Bot. 29:587–598.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, et al. 2000. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot J Linn Soc. 133:381–461.
- van Ham RCHJ. 1995. Phylogenetic relationships in the Crassulaceae inferred from chloroplast DNA variation. In: Hart H’t, Eggli U, editors. Evolution and systematics of the Crassulaceae. Leiden: Blackhuys Publishers; p. 16–29.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.