Abstract
Historically, people made a mistake on species identification on musk deer from Shanxi population. In order to further confirm the species of this population, we sequenced its mitochondrial genome and conducted the species identification. Moreover, the genetic relationships among the sister species of musk deer were also investigated. The results showed that the species of musk deer from the Shanxi population was Moschus berezovskii, and M. anhuiensis was an independent species that had a very close relationship with M. berezovskii. Additionally, M. anhuiensis and M. berezovskii might have experienced a recent history of reproductive isolation.
Musk deer of the Shanxi population had been designated Moschus moschiferus for a long time (Groves et al. Citation1995; Yang et al. Citation2003; Liao and Li Citation2008; Smith and Xie Citation2009; Wang and Zhao Citation2011). Recently, this population was reassigned to be M. berezovskii rather than M. moschiferus based on mitochondrial genes (COI and D-loop) (Yang et al. Citation2015). However, no other evidences were provided. In the present study, in order to further identify the musk deer species from the Shanxi population and investigate the genetic relationships with its sister species, we sequenced a mitochondrial genome from this population and conducted a comparison with other musk deer.
The skin from a male musk deer sample (No. XNG-md-0006) stored in the Natural History Museum of Sichuan University was collected from Xinnangou, Guandi Forest District, Shanxi province, China in 2010. We acquired its whole sequence of the mitochondrial genome according to the method described by Yang et al. (Citation2012). The mitochondrial genomic structure of musk deer from Shanxi population were determined and deposited in GenBank with the accession number JQ409122. The total length of the sequence was 16,353 bp, and its organization and gene order were similar to those of other musk deer mitochondrial genomes (Peng et al. Citation2009; Pan et al. Citation2015). The percent similarities were calculated from complete mitochondrial DNA sequences and entire amino-acid sequences (which was estimated from the nucleotide sequences) among M. anhuiensis (KP684124, NC_020017), M. berezovskii (NC_012694, JQ409122), M. chrysogaster (KC425457, KP684123), and M. moschiferus (NC_013753, JN632662) using WATER program of the EMOSS suit (Rice et al. Citation2000). The results showed that the intra-species similarities of the complete mitochondrial nucleotide sequences and amino-acid sequences were 99.2–99.9% and 99.5–99.9%, respectively. Hence, the mitochondrial nucleotide (99.2%) and amino-acid (99.8%) sequence similarities between musk deer from Shanxi population (M. berezovskii JQ409122) and M. berezovskii (NC_012694) belonged to inra-species level. The inter-species similarities of the complete/segment mitochondrial nucleotide and amino-acid sequences between M. berezovskii and M. anhuiensis were generally higher than that from other two species of musk deer.
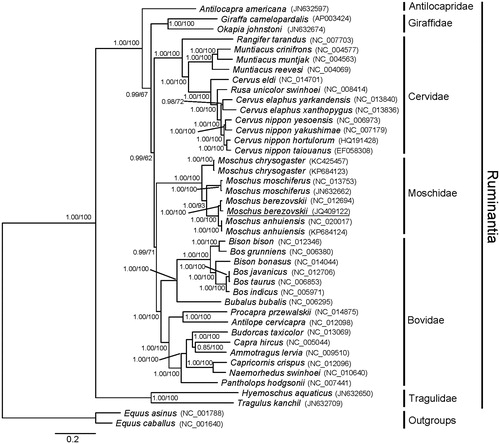
The phylogenetic trees of the Ruminantia, constructed by the BI (GTR + I + G as best fitting model) and ML methods using the nucleotide sequence data of 12 heave-strand protein-coding genes of Shanxi population of musk deer and 41 other sequences, showed the same topology (). Within Moschidae, the Shanxi population of musk deer (M. berezovskii JQ409122) clustered with M. berezovskii (NC_012694) with strong support values (PP = 1.00, BS = 100). And the high-altitude species M. chrysogaster diverged earlier than the other three species in the present study. M. berezovskii and M. anhuiensis clustered together as a clade that was sister to M. Moschiferus (). These results implied that the species of musk deer from the Shanxi population was M. berezovskii, and M. anhuiensis was an independent species that had a very close relationship with M. berezovskii. Additionally, M. anhuiensis and M. berezovskii might have experienced a recent history of reproductive isolation.
Figure 1. Molecular phylogenetic tree derived from complete DNA sequence of 12 mitochondrial protein-coding genes using Bayesian inference and ML analysis. The numbers beside the nodes are Bayesian posterior probabilities (PP) and bootstrap support (BS). Equus asinus and E. caballus were set as outgroups.
Acknowledgements
We would like to thank Mr. Guo Cai for his help in sample collection.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Groves CP, Wang Y, Grubb P. 1995. Taxonomy of musk-deer, genus Moschus (Moschidae, Mammalia). Acta Theriol Sin. 15:181–197.
- Liao X, Li Q. 2008. The geographical distribution reports of Moschus moschiferus L. in Shanxi. J Shanxi Agri Sci. 36:55–56.
- Pan T, Wang H, Hu C, Sun Z, Zhu X, Meng T, Meng X, Zhang B. 2015. Species delimitation in the genus Moschus (Ruminantia: Moschidae) and its high-plateau origin. PLoS One. 10:e0134183.
- Peng H, Liu S, Zeng B, Zou F, Zhang X, Yue B. 2009. The complete mitochondrial genome and phylogenetic analysis of forest musk deer (Moschus berezovskii). J Nat Hist. 43:1219–1227.
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the european molecular biology open software suite. Trends Genet. 16:276–277.
- Smith AT, Xie Y. 2009. A guide to the mammals of China. Changsha: Hunan Education Press.
- Wang G, Zhao Y. 2011. Survey of wildlife resources in Pangquangou. J Econ Anim. 15:134–137.
- Yang C, Li P, Zhang X, Guo Y, Gao Y, Xiong Y, Wang L, Qi W, Yue B. 2012. The complete mitochondrial genome of the Chinese Sika deer (Cervus nippon Temminck, 1838), and phylogenetic analysis among Cervidae, Moschidae and Bovidae. J Nat Hist. 46:1747–1759.
- Yang C, Xiao Z, Zou Y, Zhang X, Yang B, Hao Y, Moermond T, Yue B. 2015. DNA barcoding revises a misidentification on musk deer. Mitochondrial DNA. 26:605–612.
- Yang Q, Meng X, Xia L, Feng Z. 2003. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biol Conserv. 109:333–342.
