Abstract
We report two complete mitochondrial genome sequences of a tuber-bearing wild potato species (Solanum commersonii). The genomes are circular DNA molecules with lengths of 213,676 bp and 338,427 bp containing 80 nonredundant genes totally, including 34 protein-coding genes, 25 hypothetical open reading frames, 18 tRNA genes, and 3 rRNA genes. Phylogenetic analysis using common protein-coding sequences confirmed that S. commersonii belongs to the Solanoideae subfamily in the Solanaceae family.
Solanum commersonii is a tuber-bearing wild potato species native to Central and South America (Hawkes Citation1990). Because of its differences in several desirable characteristics from the cultivated potato (S. tuberosum) (Spooner et al. Citation1991; Rodriguez and Spooner Citation2009; Cho et al. Citation2016), the species has been used for potato breeding (Laferriere et al. Citation1999; Kim-Lee et al. Citation2005; Cho et al. Citation2016). However, the two species are sexually incompatible because of different endosperm balance numbers (EBNs) and ploidy levels of the genomes, with S. commersonii and S. tuberosum reported as diploid and tetraploid with EBN values of 1 and 4, respectively (Johnston et al. Citation1980; Ortiz and Ehlenfeldt Citation1992; Cho et al. Citation1997). Therefore, in this study, we sequenced the entire mitochondrial genome of the wild potato, S. commersonii for further investigation of its evolutionary aspects. This is the first report on a wild potato species.
Total genomic DNA was isolated from fresh leaves of the S. commersonii breeding line (specimen deposition no. Lz3.2) and used to construct a paired-end (PE) library with an insert size of ∼670 bp, according to the standard Illumina protocol for PE library construction. The library was sequenced using an Illumina MiSeq platform by LabGenomics (www.labgenomics.co.kr, Seoul, Korea), and approximately 6.8 Gb of PE reads were obtained. After removing low-quality (Q < 30) reads using CLC quality trim (ver. 4.21, CLC Inc., Denmark), approximately 4.8 Gb of high-quality reads (approximately 91× coverage for complete mitochondrial genomes) were assembled de novo with CLC genome assembler (ver. 4.21), as described previously (Cho et al. Citation2015). The longest contigs of the mitochondrial genome sequences were selected, extended, and joined via a series of PE read mapping and gap filling. The assembled mitochondrial genome sequences were corrected manually based on read mapping status and BLASTN search. The genome sequences were annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html), and manually curated based on BLAST searches.
The two complete mitochondrial genome sequences of S. commersonii, which are circular DNA molecules with lengths of 213,676 bp with a G + C content of 45.5%, and 338,427 bp with a G + C content of 45.0% were assembled independently. The two genomes showed 48.3% sequence similarity, and had two collinear regions with lengths of 126 kb and 12 kb between each other. The numbers of genes predicted were 65 and 51, respectively, in the two genomes, constituting approximately 12.8% and 13.75%, respectively, the two genome sequences. Among the predicted genes, 10 protein-coding genes, 14 hypothetical open reading frames (ORFs), 2 rRNA genes, and 8 tRNA genes were commonly present between the two genomes. Overall, 80 unique genes, including 34 protein-coding genes, 25 ORFs, 18 tRNA genes, and 3 rRNA genes were identified in the genomes. The exons of the trans-spliced genes, nad1, nad2, and nad5, exist completely or partially in each genome.
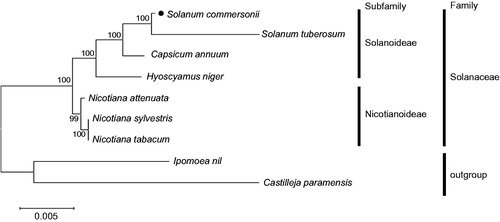
Phylogenetic analysis was performed based on multiple alignments of common 23 protein-coding sequences in mitochondrial genome, and confirmed that S. commersonii belongs to the Solanoideae subfamily in the Solanaceae family ().
Figure 1. Maximum likelihood (ML) tree of nine Solanaceae-related species. The 23 protein-coding sequences in mitochondrial common gene were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html). And then, phylogenetic tree was constructed with tamura-nei model and 1000 bootstrap method by MEGA 7.0 (Kumar et al. Citation2016). Mitochondrial genome sequences used for this tree are Capsicum annuum, NC_024624; Castilleja paramensis, NC_031806; Hyoscyamus niger, NC_026515; Ipomoea nil, NC_031158; Nicotiana attenuate, MF579563; N. sylvestris, NC_029805; N. tabacum, NC_006581; S. commersonii, MF989960 and MF989961; S. tuberosum, MF989953 to MF989957.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cho K-S, Cheon K-S, Hong S-Y, Cho J-H, Im J-S, Mekapogu M, Yu Y-S, Park T-H. 2016. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Rep. 35:2113–2123.
- Cho HM, Kim-Lee HY, Om YH, Kim JK. 1997. Influence of endosperm balance number (EBN) in interploidal and interspecific crosses between Solanum tuberosum dihaploids and wild species. Korean J Breed. 29:154–161.
- Cho K-S, Yun B-K, Yoon Y-H, Hong S-Y, Mekapogu M, Kim K-H, Yang T-J. 2015. Complete chloroplast genome sequence of tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLoS One. 10:e0125332.
- Hawkes JG. 1990. The potato: evolution, biodiversity and genetic resources. London (UK): Belhaven Press.
- Johnston SA, den Nijs TP, Peloquin SJ, Hanneman RE. Jr. 1980. The significance of genic balance to endosperm development in interspecific crosses. Thero Appl Genet. 57:5–9.
- Kim-Lee HY, Moon JS, Hong YJ, Kim MS, Cho HM. 2005. Bacterial wilt resistance in the progenies of the fusion hybrids between haploid of potato and Solanum commersonii. Am J Potato Res. 82:129–137.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laferriere LT, Helgeson JP, Allen C. 1999. Fertile Solanum tuberosum + S. commersonii somatic hybrids as sources of resistance to bacterial wilt caused by Ralstonia solanacearum. Theor Appl Genet. 98:1272–1278.
- Ortiz R, Ehlenfeldt MK. 1992. The importance of endosperm balance number in potato breeding and the evolution of tuber-bearing Solanum species. Euphytica. 60:105–113.
- Rodriguez F, Spooner DM. 2009. Nitrate reductase phylogeny of potato (Solanum sect. Petota) genomes with emphasis on the origins of the polyploid species. Syst Bot. 34:207–219.
- Spooner DM, Bamberg J, Hjerting JP, Gomez J. 1991. Mexico, 1988 Potato germplasm collecting expedition and utility of the Mexican potato species. Am Potato J. 68:29–43.
