Abstract
Prunus takesimensis is an endemic flowering cherry on Ulleung Island, Korea. The complete chloroplast (cp) genome of P. takesimensis was generated by de novo assembly using whole-genome next-generation sequencing approach. The cp genome size was 157,948 bp in length consisting of four regions; large single copy region (85,959 bp), small single copy region (19,117 bp), and a pair of inverted repeat regions (26,436 bp). The genome contained a total of 128 genes, including 83 coding genes, 8 rRNA genes, and 37 tRNA genes. Phylogenetic analysis for 20 reported genomes within the family Rosaceae showed the monophyly of the genus Prunus including newly sequenced P. takesimensis.
The wild flowering cherry, Prunus takesimensis Nakai is distributed on Ulleung Island, Korea. Morphologically, P. takesimensis is the most similar to P. sargentii, which is found in the adjacent continent and islands (i.e. Jeju Island) of Korea, Japanese Archipelago, and Russian Far East (Ohwi Citation1965; Kim Citation2009). Due to the morphological similarities shared by both species, P. sargentii is presumed as the continental progenitor of P. takesimensis. The Ulleung Island endemic P. takesimensis has been recognized from its sister species primarily based on reproductive traits: smaller flowers (26–32 mm in diameter) and more flowered (3–5) inflorescence (Chang et al. Citation2004). However, the genetic relationship of P. takesimensis relative to other flowering cherries has not been well established in the previous phylogenetic analyses using several nuclear and chloroplast markers (Cho Citation2017). Therefore, we sequenced the whole chloroplast genome of P. takesimensis to elucidate its evolutionary history on Ulleung Island in the context of the phylogenetic relationship among other flowering cherries.
Total genomic DNA was isolated from fresh leaves sampled from natural habitat (37°29′3.83″N, 130°54′44.37″E, Alt. 132m) in Ulleung Island using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA). Voucher specimen (SKK Cho et al. PT1) was deposited in the herbarium of Sungkyunkwan University (SKK). Paired-end genomic library was constructed and sequenced using Illumina HiSeq 4000 platform. Total reads of 10.95 Gb (Order 1611AHF-0002/sample PT1) were obtained and assembled by de novo genomic assembler, Velvet 1.2.10 (Zerbino and Birney Citation2008). The representative chloroplast contig (>521×) was retrieved and aligned against the reference chloroplast genome of P. serrulata var. spontanea (GenBank accession KP760073) using Geneious v8.1.6 (Biomatters Ltd., Auckland, New Zealand). Annotation was performed using Dual Organellar GenoMe Annotator (Wyman et al. Citation2004), ARAGORN v1.2.36 (Laslett and Canback Citation2004) and RNAmmer 1.2 Server (Lagesen et al. Citation2007).
The complete chloroplast genome of P. takesimensis (GenBank accession MG754959) was 157,948 bp in length consisting of four regions; large single copy region (LSC) of 85,959 bp, small single copy region (SSC) of 19,117 bp, and a pair of inverted repeat regions (IRA and IRB) of each 26,436 bp. The overall GC contents of the chloroplast genome were 36.7%; LSC (34.6%), SSC (30.2%), and IR (42.5%) regions. The genome contained a total of 128 genes, including 83 coding genes, 8 rRNA genes, and 37 tRNA genes.
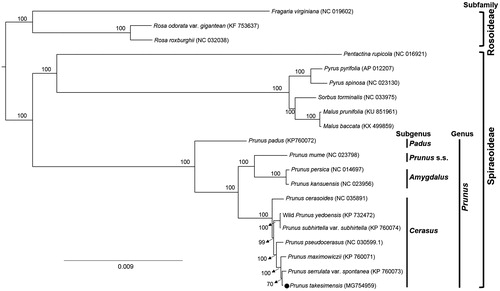
Phylogenetic analysis was conducted to confirm the phylogenetic position of newly sequenced P. takesimensis amongst 20 representative species of Rosaceae. The complete chloroplast genome sequences were aligned using MAFFT v.7 (Katoh and Standley Citation2013) to produce the maximum likelihood (ML) tree by IQ-TREE (Nguyen et al. Citation2014) with 1000 replicate bootstrap (BS) analyses. The ML tree () showed that the genus Prunus was strongly supported as monophyletic group (BS 100%) and P. takesimensis was most closely related to P. serrulata var. spontanea.
Figure 1. Maximum likelihood tree of the Rosaceae species constructed by IQ-TREE. Twenty representative complete chloroplast genome sequences were used in the analysis including P. takesimensis (GenBank accession MG754959). The percentage of bootstrap support value calculated with 1000 bootstrap replications is shown on each node. The phylogenetic tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Disclosure statement
The authors report no potential conflict of interest. The authors are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chang CS, Choi H, Chang KS. 2004. Reconsideration of Prunus sargentii complex in Korea – with respect to P. sargentii and P. takesimensis. Korean J Pl Taxon. 34:221–244. (in Korean).
- Cho MS. 2017. The origin and evolution of the flowering cherries (Prunus; Rosaceae), endemic to the volcanic islands in Korea [dissertation]. Suwon, Korea: Sungkyunkwan University.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim CS. 2009. Vascular plant diversity of Jeju Island, Korea. Korean J Plant Res. 22:558–570.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 35:3100–3108.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2014. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ohwi J. 1965. Flora of Japan. Washington (DC): Smithsonian Institution.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
