Abstract
The complete mitochondrial genome is sequenced and analyzed from a xanthid crab Etisus anaglyptus, which is the first complete mitochondrial genome for the genus. The mitochondrial genome length of E. anaglyptus is 16,435 bp and it is composed of 13 protein coding genes, two ribosomal RNA genes, and 22 tRNA genes. The structure and gene orientation of the mitochondrial genome is identical with the other brachyuran records. Furthermore, phylogenetic relationships of the infraorder Brachyura evaluated by mitochondrial protein coding genes. The phylogenetic study showed that E. anaglyptus is positioned in the superfamily Xanthoidea and the closest species to E. anaglyptus is Leptodius sanguineus.
Xanthidae is one of the most dominant crab family in the brachyuran with 572 species in 133 genera (De Grave et al. Citation2009). Although the morphologic identification of xanthids is well studied, mitochondrial genome records of their species are inadequate. There are only one complete mitochondrial genome recorded (Sung et al. Citation2016). In the present study, we are providing complete mitochondrial genome analysis of a Xanthidae species Etisus anaglyptus. Besides, due to the complete mitochondrial genome information, phylogenetic relationship of the species in the superfamily Xanthoidea is investigated. This is the second record for the family Xanthidae and the first record for the genus Etisus.
The species have been collected from the rocky intertidal zone of Con Co Island, Vietnam (17° 9′ 54″ N, 107° 20′ 40″ E) on July 2017 and preserved in 97% ethanol. Whole genomic DNA was extracted from walking legs. The methods for sequencing and analyzing complete mitochondrial genome were described previously (Karagozlu et al. Citation2016). The specimen stored in Department of Biotechnology, Sangmyung University, Korea (SM00247).
Mitochondrial genome length of E. anaglyptus is 16,435 bp (GenBank accession number: MG751773). It is composed of 13 protein coding genes, two ribosomal RNA genes, and 22 tRNA genes. The structure and gene orientation of the mitochondrial genome is typical to brachyuran species (Miller et al. Citation2004). Likewise, all brachyuran records 23 genes encoded on the majority strand and 14 genes positioned in minority strand. The nucleotide distribution of the mitochondrial genome is 33.2% A, 21% C, 11.1% G, and 34.7% T. There are 15 overlapping regions with 1–25 bp in length. The longest overlapping region is located between nad1 and tRNALeu. Besides, the mitochondrial genome has 16 intergenic sequences varying from 1 to 633 bp in length. The largest intergenic sequence is located between 16S rRNA and tRNAVal. In the mitochondrial genome the genes are using three different initiation codons which are ATA, ATG, and ATT. The most common starting codon is ATG that used by cytb, cox1, cox2, cox3, atp8, nad4, and nad4l genes. Also, there are two different stop codons observed which are TAA and TAG.
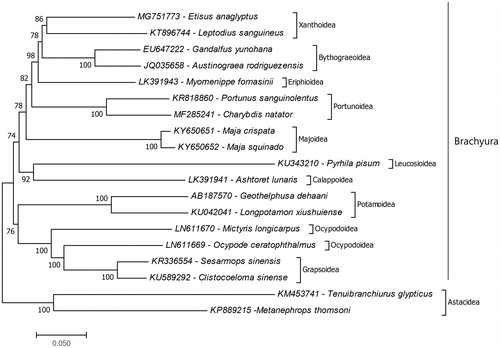
The phylogenetic study showed that E. anaglyptus positioned in the superfamily Xanthoidea and the closest species to E. anaglyptus is Leptodius sanguineus (KT896744) which is the first complete mitochondrial genome record from the family Xanthidae (). Also, the superfamily Xanthoidea has sister group relationship with the superfamily Bythograeoidea. Similar results showed previously by combination of the six nuclear genes and two mitochondrial genes based molecular study (Tsang et al. Citation2014) and protein coding genes of complete mitochondrial genome based study (Sung et al. Citation2016). Previously, the genus has been shown to be polyphyletic by three mitochondrial and one nuclear marker based molecular phylogeny study (Lai et al. Citation2011). Unfortunately, there is no enough mitochondrial genome data recorded to confirm their relationship additional complete mitochondrial genome data from the genus should provide. The present study presents additional data for molecular Xanthidae phylogeny.
Figure 1. Phylogenetic relationships of that Etisus anaglyptus in the infraorder Brachyura due to amino acid sequences of mitochondrial protein coding genes. The mitochondrial genome data which belong to superfamilies of the infraorder Brachyura retrieved from GenBank. Maximum two species chosen as representative of brachyuran super families and the species belongs to the infraorder Astacidea chosen as representative of outgroup. For reconstruction of the phylogenetic tree maximum likelihood statistical method used and bootstrap method replicated 1000 times for the test of phylogeny.
Acknowledgements
We thank the Institute of Marine Environment and Resources (IMER), for allowing marine organism research.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- De Grave S, Pentcheff ND, Ahyong ST, Chan TY, Crandall KA, Dworschak PC, Felder DL, Feldmann RM, Fransen CHJM, Goulding LYD, et al. 2009. A classification of living and fossil genera of decapod crustaceans. Raff Bull Zool Suppl. 21:1–09.
- Karagozlu MZ, Sung JM, Lee JH, Kwon T, Kim CB. 2016. Complete mitochondrial genome sequences and phylogenetic relationship of Elysia ornata (Swainson, 1840) (Mollusca, Gastropoda, Heterobranchia, Sacoglossa). Mitochondrial DNA Part B. 1:230–232.
- Lai JCY, Mendoza JCE, Guinot D, Clark PF, Ng PKL. 2011. Xanthidae MacLeay, 1838 (Decapoda: Brachyura: Xanthoidea) systematics: a multi-gene approach with support from adult and zoeal morphology. Zool Anz. 250:407–448.
- Miller AD, Nguyen TTT, Burridge CP, Austin CM. 2004. Complete mitochondrial DNA sequence of the Australian freshwater crayfish, Cherax destructor (Crustacea: Decapoda: Parastacidae): a novel gene order revealed. Gene. 331:65–72.
- Sung JM, Lee JH, Kim SG, Karagozlu MZ, Kim CB. 2016. Complete mitochondrial genome of Leptodius sanguineus (Decapoda, Xanthidae). Mitochondrial DNA Part B. 1:500–501.
- Tsang LM, Schubart CD, Ahyong ST, Lai JCY, Au EYC, Chan TY, Ng PKL, Chu KH. 2014. Evolutionary history of true crabs (Crustacea: Decapoda: Brachyura) and the origins of freshwater crabs. Mol Biol Evol. 31:1173–1187.
